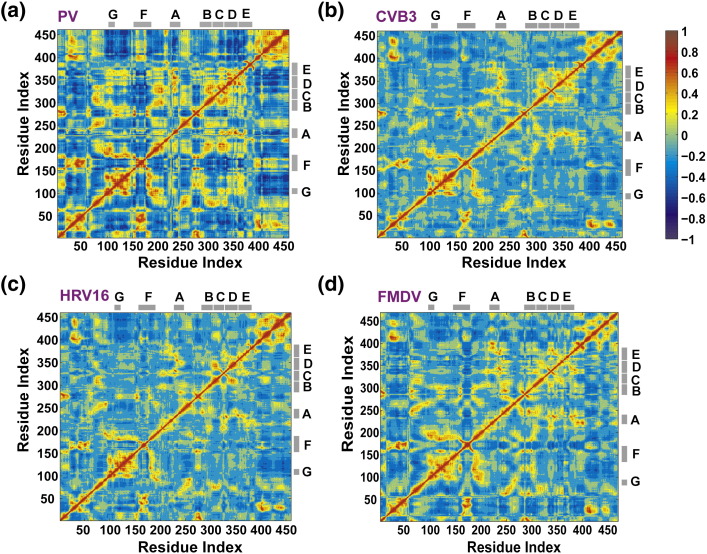
Fig. 5.
Correlated motions in RdRps. The DCCM, which measures the correlation between the displacements of Cα atoms, calculated from simulations of RdRps from PV (a), CVB3 (b), HRV16 (c) and FMDV (d) is shown. For each enzyme, the calculated matrix whose elements are the pairwise correlation scores between its residues is visualized as a colored map. The correlation scores are encoded with a color gradient from − 1 (blue, completely anti-correlated) to + 1 (red, completely correlated). The conserved structural motifs are marked with gray bars. Residues that appeared flexible in PCA of PV RdRp (see Fig. 4a) are shown to be strongly correlated. Both positive and negative correlations have implications regarding polymerase functions as discussed in the text. The figure was prepared using MATLAB 7.6.