Fig. 6.
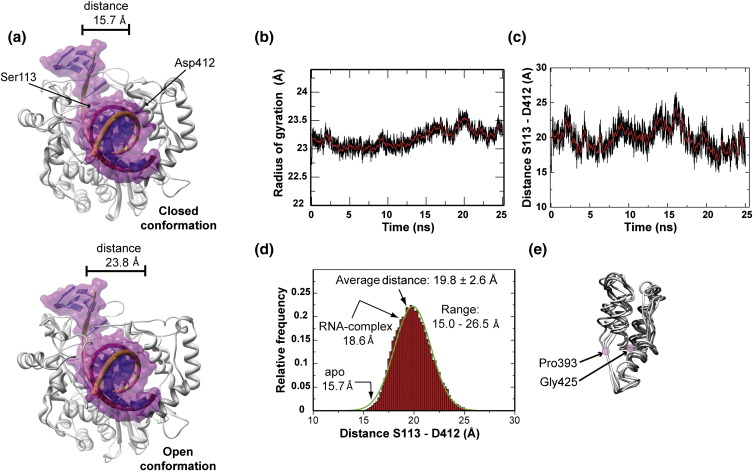
Opening and closing of the RNA-binding channel of PV RdRp. (a) The closed (top) and open (bottom) conformations of PV RdRp with modeled RNA. The RNA modeled in the apoenzyme representing the closed conformation was taken from the PV RdRp elongation complex structure (PDB code 3OL6). The open conformation derives from a snapshot from the simulation. (b) Oscillation of the radius of gyration (Rg). The time evolution of Rg for the simulated structure is shown by the black line; the running average (200-ps window) is shown by the red line. (c) Oscillation of the Cα–Cα distance of Ser113 (fingers) and Asp412 (thumb). The time evolution of distance for the simulated structure is shown by the black line; the running average (200-ps window) is shown by the red line. (d) The frequency distribution of the distances observed in (c). The green line represents the fit to a Gaussian model using the program Grace-5.1.22. (e) Hinge-twist motion of the thumb observed by superimposition of snapshots across the simulation trajectory. Residues P393 and G425 are the pivot points for the motion.