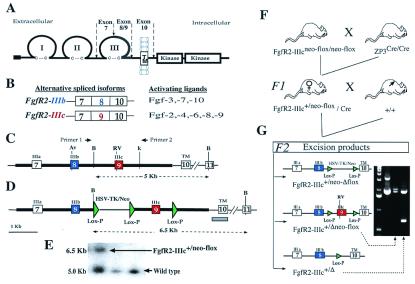
Figure 1.
Fgf receptor structure and strategy for deletion of FgfR2 exon 9 (IIIc). (A) Schematic structure of FgfR2 highlighting the third Ig loop, a region in which alternative usage of exons 8 or 9 in FgfR2 leads to the generation of the IIIb or IIIc isoforms, respectively. TM, transmembrane domain. (B) The main ligands activating each of these isoforms. (C) Schematic depiction of mouse genomic DNA encompassing FgfR2 exons (boxes) 7, 8, 9, and 10, drawn to scale and showing main restriction enzyme sites (Av, AvaI; RV, EcoRV; B, BamHI; K, KpnI). (D) Targeting construct showing loxP sequences placed downstream of exon 9, and flanking the selectable marker gene neo driven by HSV-TK promoter located upstream of exon 9. Thick lines indicate the extent of the targeting construct. (E) Homologous recombinant 129 embryonic stem cells were identified by Southern blotting of BamHI-digested DNA by using a 450-bp genomic probe located 3′ of target vector sequences (gray bar in D). Homologous recombinant cells (FgfR2-IIIc+/neo-flox) yielded a 6.5-kb fragment and wild type yielded a 5.0-kb fragment. Both 5′ and 3′ joins were checked by PCR analysis. (F) Hemizygous (FgfR2-IIIc+/Δ) mutant mice were generated by crossing FgfR2-IIIcneo-flox/neo-flox with ZP3-Cre females (34), and then crossing F1 females carrying one copy of the targeting construct as well as the Cre transgene with wild-type males. Approximately 50% of such females gave Cre-mediated excision. (G) Of these excisions, 90% were complete (FgfR2-IIIc+/Δ); the remainder excised only the selectable marker (FgfR2-IIIc+/Δneo-flox), as determined by PCR using the pair of primers shown in C. The potential excision product, FgfR2-IIIc+/neo-Δflox, was not observed.