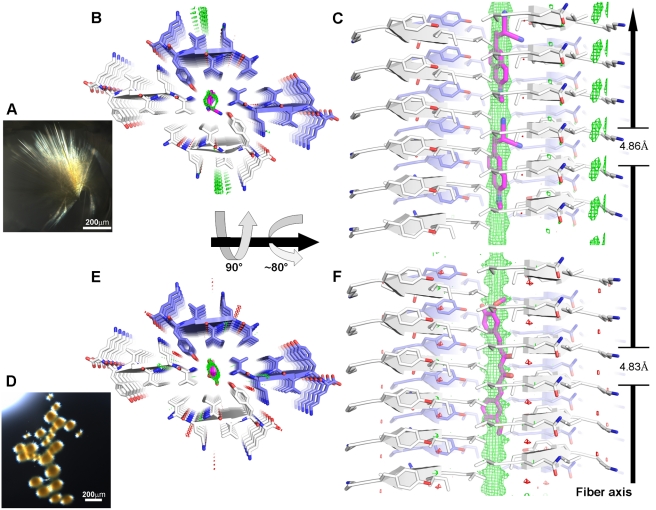
Figure 6. Models of DDNP and curcumin bound to the VQIVYK fiber based on undifferentiated electron density.
Panels A and D are micro-crystals of VQIVYK co-crystallized with DDNP and curcumin, respectively. In the structure of the complexes with DDNP (B–C) and curcumin (E–F), VQIVYK is packed in a form having a steric zipper with one β-sheet shifted in relation to the other β-sheet (cartoon arrows). The carbons of VQIVYK are colored white for one steric zipper and blue for the other. VQIVYK, DDNP, and curcumin are shown as sticks with non-carbon atoms colored by atom type. Six layers of the fiber are depicted. In panels B and E, the view looks down the fiber axis. In panels C and F, the view is perpendicular to the fiber axis. In both complexes, only the VQIVYK segment is modeled into the electron density, and in both, there is an apparent difference electron density Fo-Fc map (shown as mesh, +3σ in green and −3σ in red) located in the void formed by the shift of the steric zipper. The positive density (part of the structure that has not been modeled, green mesh) displays a continuous tube-like shape, running along the fiber axis. We attribute this density to the presence of the small molecules, yet it is too undifferentiated to fit atoms in detail. DDNP (B–C, two molecules are shown) and curcumin (E–F) (both in magenta carbons) have been computationally docked (Methods) into the structures and fit the location of the positive density. The unit cell dimension of the crystal along the fiber axis is indicated. The length of both DDNP and curcumin spans multiple unit cells of the fibril; that is, the dimensions of the small molecule and the fibril unit cell are incommensurate (see Text S1).