Abstract
The presence of externally supplied DNA in the growth medium enhances growth of lateral roots and root hairs in Arabidopsis. This phenomenon cannot be attributed to phosphorus (P) limitation because it is independent of the plants' P status. Rather, we hypothesized that DNA triggers a currently unknown signaling pathway. Analyzing the transcriptional changes of genes induced by externally supplied DNA, we show that 7 of the 17 studied CLAVATA3/ESR-related (CLEs) genes were differentially regulated. CLEs are known peptide hormones that affect plant development including root morphology. While previous research had shown that overexpression of these CLE genes alters root morphology, changes in gene expression had not been linked to environmental triggers. The differential expression of these CLEs genes and accompanied changes of the root phenotype are indicative of a DNA-elicited signal pathway which affects root development. We conclude that DNA acts as a signaling compound which induces root proliferation in a way that would enhance the plant's ability to acquire nutrients from soil organic matter. Our study further confirms the importance of CLEs for controlling root morphology in response to specific environmental conditions, and draws attention to a novel role of DNA as a signaling compound.
Key words: arabidopsis, DNA, CLEs, root morphology, plant signaling
New evidence of how plants acquire nutrients has emerged indicating that plants do not only rely on small molecules or ions to supply mineral nutrients for growth. We recently showed that plants are able to incorporate large organic molecules including proteins and DNA into roots.1,2 This contradicts the notion that large organic molecules become available for plant use only after microbial conversion into small molecules and ions.3–5 In Arabidopsis, supply of protein (bovine serum albumin) in the growth medium could only partially compensate for the lack of inorganic nitrogen in the growth medium.1 In contrast, DNA fully substituted for the absence of inorganic P (Pi) in the growth medium,2,6 implying that Arabidopsis is well adapted to use DNA as an alternative source of P for growth.
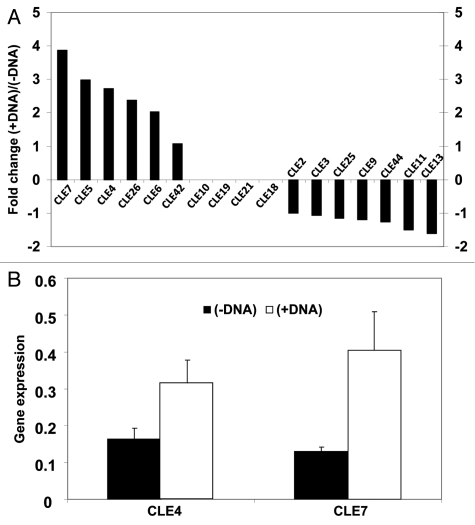
Surprisingly, the presence of DNA in Pi-containing growth medium also enhanced growth of lateral roots and root hairs although plants were P replete,2 suggesting that DNA or its degradation products act as signaling molecules that trigger changes in root morphology.2 To investigate the hypothesis that DNA is a signaling compound that affects root morphology, we analyzed gene expression of plants grown with and without DNA in the growth medium. Arabidopsis plants, grown in axenic hydroponic culture for 3 weeks with Pi supply (5.7 µg KH2PO4 per mL medium) were incubated for a further 24 h without (−DNA) or with (+DNA, 0.8 mg herring sperm DNA per mL). RNA was extracted from roots to probe an Agilent microarray (Agilent Technologies, USA). Comparative analysis of gene expression between treatments revealed that DNA addition induced changes in expression of CLAVATA3/ESR-related (CLE) genes which have a known role in root morphogenesis (Fig. 1A). Microarray data were validated by quantitative real-time PCR analysis of CLE4 and CLE7 expression as representative CLE genes with strongly increased expression (Fig. 1B).
Figure 1.
Regulation of CLE genes expression in Arabidopsis by DNA in the growth medium. (A) Microarray results showing that CLE genes are differentially regulated in the presence of DNA. Microarray data are means of two technical replicates. (B) Quantitative real-time PCR (QRT-PCR) analysis of CLE4 and CLE7 expression in the presence of DNA of Arabidopsis grown in the same condition as for the microarray. QRT-PCR showed 2-fold and 3.1-fold upregulation of CLE4 and CLE7 respectively, confirming the microarray results which show 2.7-fold and 3.9-fold upregulation, respectively. QRT-PCR results are means (± SD) of three independent biological assays and each value is the mean of three technical preparations. Expression levels were normalized using β-actin.
CLE genes encode a family of at least 32 peptide ligands involved in plant developmental processes including regulation of root and shoot apical meristems.7–9 Overexpression of certain CLE genes increases or shortens the primary root length of Arabidopsis seedling.10 The observed changes in the expression of CLE genes (Fig. 1A) may therefore be responsible for, or contribute to, the changes in root morphology observed when DNA is present in the growth medium.2
The five CLE genes which were significantly upregulated in the +DNA treatment (CLE7, AT2G31082; CLE5, AT2G31083; CLE4, AT2G31081; CLE26, AT1G69970; and CLE6, AT2G31085; Fig. 1A) induced longer primary roots in Arabidopsis seedling when overexpressed.10 In contrast, CLE13 (AT1G73965) and CLE11 (AT1G49005) that were significantly downregulated in the +DNA treatment (Fig. 1A) caused shorter primary roots in Arabidopsis seedlings when overexpressed.10 Strabala et al. (2006) reported only the effect of overexpression of CLE genes on primary root length of seedlings, while we observed a major stimulatory effect of externally supplied DNA on lateral root number, lateral root length, and root hair length in mature Arabidopsis plants.2 Nonetheless, our microarray expression data and the findings of Strabala et al. (2006) are consistent with the notion that changes in CLE genes expression mediate all or some of the effects caused by externally supplied DNA on root morphology.
Our results add important knowledge to the discussion of the role of CLE genes for plant development. A recurring issue in the study of CLE genes is the lack of phenotypes associated with mutations of individual CLE genes.9 Apart from the possible redundancy among the CLE genes, a proposed explanation is that knock-out-phenotypes are only visible under certain environmental conditions.9 Our findings strongly support this possibility as they demonstrate that a set of CLEs is differentially regulated in response to the environmental stimulus exerted by DNA and results in a co-ordinated alteration of root morphology. Therefore, plants grown in the presence of DNA may offer a model system to study the role of individual CLEs genes.
In conclusion, our results indicate that in Arabidopsis a series of CLE genes control root morphogenesis under specific environmental conditions. It is tempting to argue that the high number of possible combinations of the 32 CLEs allow plants to respond to specific growth conditions, such as different nutrient supply situations in soil. Our results are also relevant for the understanding of how plants control root growth in soils that contain organic forms of nutrients. The complex interface between plants, microbes and soil is not well understood and plants may have evolved specific pathways that allow controlling organ-morphogenesis in response to the presence of different forms of mineral nutrients.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/12477
References
- 1.Paungfoo-Lonhienne C, Lonhienne TG, Rentsch D, Robinson N, Christie M, Webb RI, et al. Plants can use protein as a nitrogen source without assistance from other organisms. Proc Natl Acad Sci USA. 2008;105:3–5. doi: 10.1073/pnas.0712078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paungfoo-Lonhienne C, Lonhienne TG, Mudge SR, Schenk PM, Christie M, Carroll BJ, et al. DNA is taken up by root hairs and pollen, and stimulates root and pollen tube growth. Plant Physiol. 2010;153:799–805. doi: 10.1104/pp.110.154963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan JA. Ecology. Looking beneath the surface. Science. 2002;298:1903–1904. doi: 10.1126/science.1079808. [DOI] [PubMed] [Google Scholar]
- 4.Bonkowski M. Protozoa and plant growth: the microbial loop in soil revisited. New Phytol. 2004;162:617–631. doi: 10.1111/j.1469-8137.2004.01066.x. [DOI] [PubMed] [Google Scholar]
- 5.Clarholm M. Interactions of Bacteria, Protozoa and Plants Leading to Mineralization of Soil-Nitrogen. Soil Biol Biochem. 1985;17:181–187. [Google Scholar]
- 6.Chen DL, Delatorre CA, Bakker A, Abel S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta. 2000;211:13–22. doi: 10.1007/s004250000271. [DOI] [PubMed] [Google Scholar]
- 7.Strabala TJ. CLE genes in plant development: Gain-of-function analyses, pleiotropy, hypermorphy and neomorphy. Plant Signal Behav. 2008;3:457–459. doi: 10.4161/psb.3.7.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miwa H, Kinoshita A, Fukuda H, Sawa S. Plant meristems: CLAVATA3/ESR-related signaling in the shoot apical meristem and the root apical meristem. J Plant Res. 2009;122:31–39. doi: 10.1007/s10265-008-0207-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Fiers M. CLE peptide signaling during plant development. Protoplasma. 2004;240:33–43. doi: 10.1007/s00709-009-0095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strabala TJ, O'Donnell PJ, Smit AM, Ampomah-Dwamena C, Martin EJ, Netzler N, et al. Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 2006;140:1331–1344. doi: 10.1104/pp.105.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]