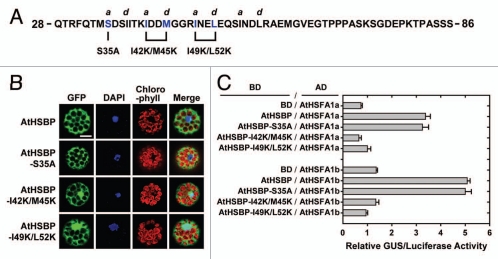
Figure 2.
AtHSBP mutagenesis, cellular localization and interaction with AtHSFs. (A) The partial protein sequence of AtHSBP, residues 28 to 86. The hallmark of coiled-coil structure is a heptad repeat of 7 amino acid residues (“abcdefg”), with a predominance of hydrophobic residues at a and d positions, and residues at e and g positions are frequently charged, as predicted by COILS (http://www.ch.embnet.org/software/COILS_form.html) in a 14-residue window. The AtHSBP-S35A, AtHSBP-I42K/M45K and AtHSBP-I49K/L52K are shown below the sequence. (B) AtHSBP and the mutations were transiently expressed in Arabidopsis mesophyll protoplasts under normal growth conditions. Green fluorescent protein (GFP) signals were observed on confocal microscopy. Blue shows the nucleus stained with 4,6-diamino-phenylindole (DAPI), and red shows the chlorophyll with auto-fluorescence. Similar results were obtained from 3 independent replicates, and representative images are shown. Bar = 20 µm. (C) Quantification of protein-protein interaction by P2H assay. AtHSBP and the mutations were fused with GAL4 DNA-binding domain (BD) or -activation domain (AD), respectively, and used for protoplast transfections, as indicated. Transfections with BD and AtHSFs-AD constructs were used as references. Interactions between AtHSBP and AtHSFs were used as positive controls. The amount of relative GUS activity was normalized by luciferase luminescence. Data are means ± SD (n = 3).