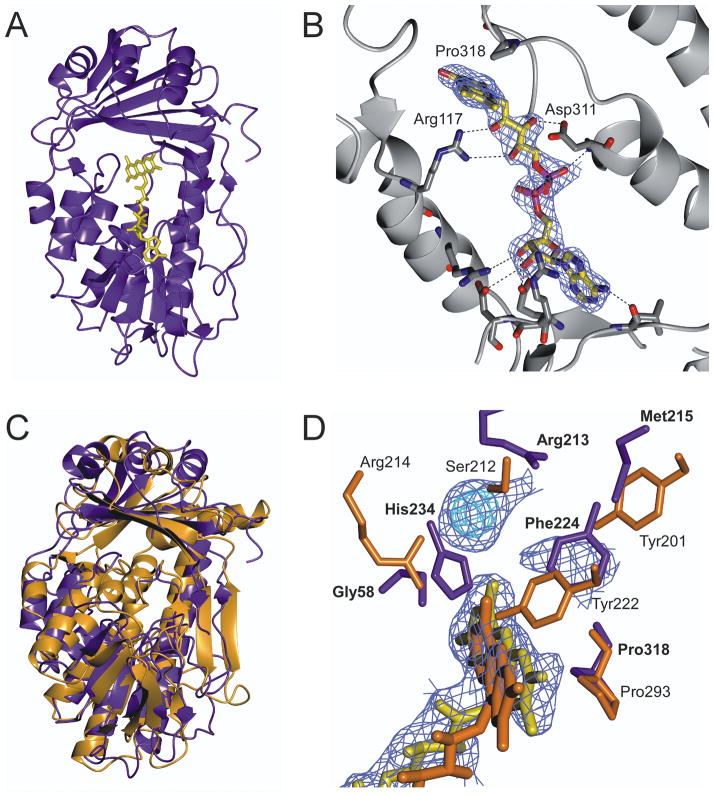
Fig. 2. Crystal structure of wild-type TetX2 from Bacteroides thetaiotaomicron, a FAD-dependent monooxygenase determined at 2.8 Å.
A) Ribbon representation of crystal structure of TetX2 with bound FAD (yellow). B) FAD binding is coordinated through a number of hydrogen interactions with residues surrounding the FAD-binding pocket. C) Superposition of structures of TetX2 (blue) and PHBH (orange) results in 4.1 Å r.m.s.d. The overall topology of TetX2 is similar to PHBH and other FAD-dependent hydroxylases, forming two domains with one FAD molecule and of the absence an independent NADHP binding domain. D) Overlay of active sites of TetX2 (blue residues) and PHBH (orange residues) with strong unknown density shown in blue and cyan. Residues surrounding the active site are not well conserved with the exception of Pro318 (Pro293 in PHBH). Final (2Fo−Fc) SIGMAA weighted electron density map corresponding to FAD and the unknown density was contoured at 1.5σ (blue) and 3σ (cyan). Figure was made using CCP4mg20.