Figure 1. L. monocytogenes PrsA2 domain organization and construction of PrsA1/PrsA2 domain swap mutants.
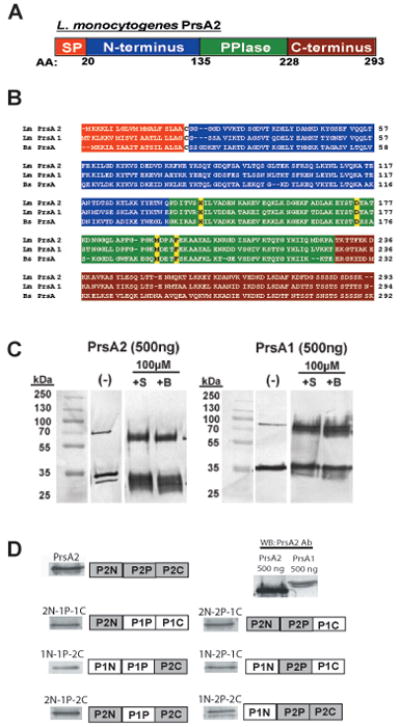
(A) Illustration of the predicted domain architecture of PrsA2. Included are an N-terminal signal peptide (orange), an N-terminal domain of 135 amino acids (blue), a central PPIase domain of 93 amino acids (green), and a C-terminal domain of 66 amino acids (red). (B) Chemical crosslinking of purified PrsA1 and PrsA2. 500ng of purified PrsA1 and PrsA2 proteins were chemically crosslinked with 10μM of sulfo-ethylene glycol bis[succinimidylsuccinate] (+S) or Bis[sulfosuccinimidyl] suberate (+B) for 1 hour at room-temperature followed by SDS-PAGE and western blotting for detection of PrsA dimers. All data is representative of at least three independent experiments. (−), no crosslinker added. (C) Amino acid sequence alignment of L. monocytogenes PrsA2 and PrsA1 and B. subtilis PrsA. Each domain is colored as in (A). The four amino acids known to be critical for PPIase activity in B. subtilis PrsA are highlighted in yellow. (D) PrsA1/PrsA2 chimeric domain swap mutants introduced into ΔprsA1 ΔprsA2 mutant strains using the site-specific integration vector pPL2. Each construct is under the expression control of the prsA2 promoter. The six constructs generated are shown along with respective Western blots using PrsA2 polyclonal antibody. Because all of the constructs generated contain a substantial portion of the PrsA1 protein, a Western blot was also performed on equivalent amounts (500 ng) of PrsA1 and PrsA2 using polyclonal PrsA2 antibody. Based on differences in antibody recognition of PrsA1 and PrsA2, approximately equivalent amounts of chimera proteins were expressed in L. monocytogenes (see also Supplementary Fig. S1).
