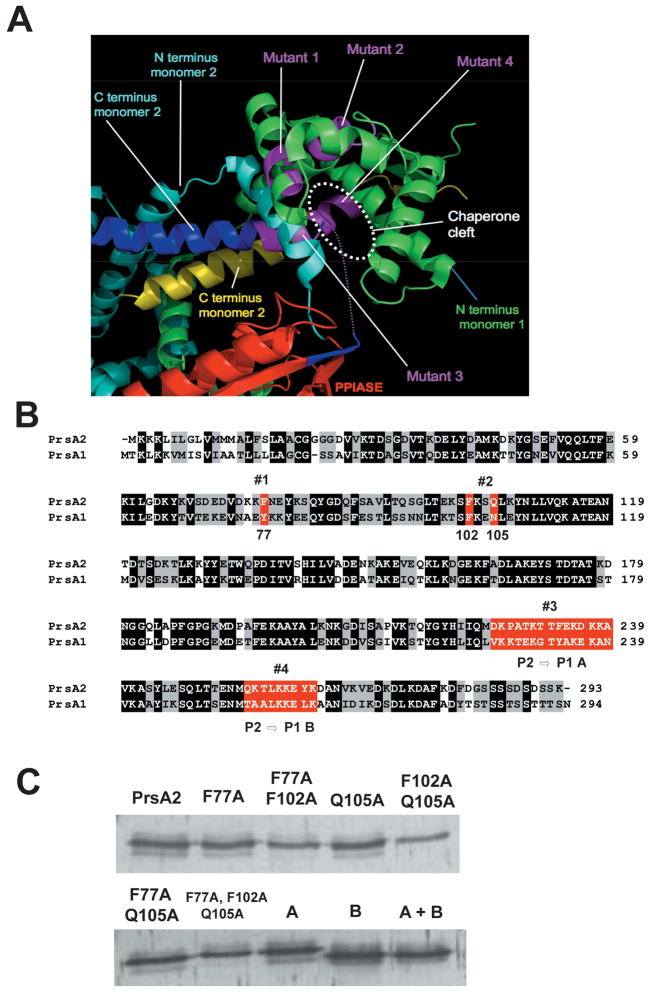
Figure 2. The structure of E. coli SurA indicating the position of mutations introduced in this study.
(A) The structure of SurA was used as a guide to design mutations in PrsA2. By analogy with SurA, PrsA2 is predicted to act as a dimer with contributions from both the N and C-terminus of each monomer. PPIase domain (red), N-terminus of monomer 1 (green), N-terminus of monomer 2 (light blue), C terminus of monomer 2 (yellow and blue). Purple regions represent predicted contact regions for substrate binding in the C-terminus of monomer 2 and the N-terminus of monomer 1. (B) Amino acid sequence alignment of PrsA2 and PrsA1. Black shading indicates identical amino acids between PrsA2 and PrsA1; gray shading indicates similar amino acids; and red highlighting represents the four initial regions of the putative substrate binding cleft targeted for mutagenesis. Western blot of the nine mutants is shown (C).