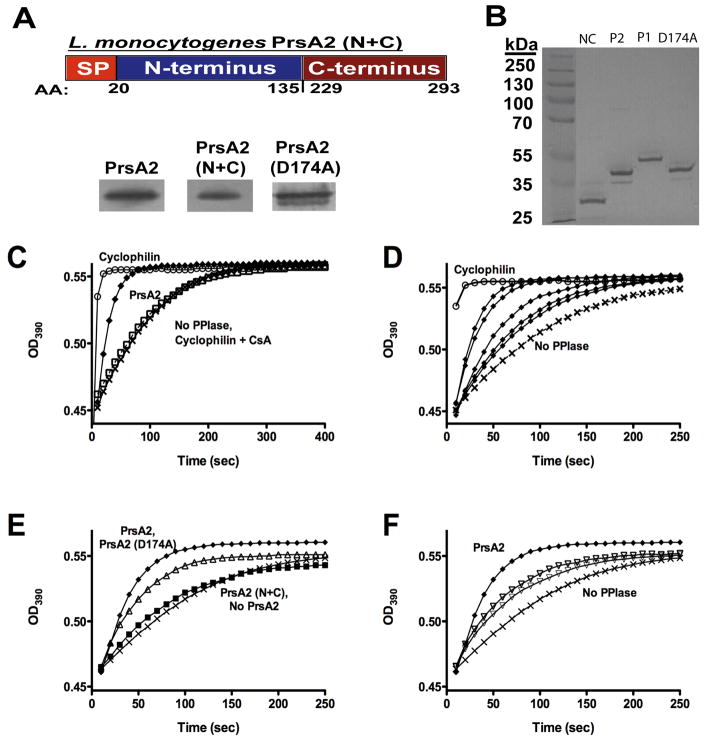
Figure 3. The central domain of PrsA2 has PPIase activity.
(A) Upper panel: construction of a PrsA2 N+C mutant lacking the entire central PPIase domain. Lower panels: PrsA2 Western blots of ΔprsA2 + pPL2- prsA2, ΔprsA2 + pPL2-prsA2 N+C, andΔprsA2 + pPL2-prsA2 D174A surface proteins to confirm synthesis and localization of PrsA2 mutant constructs. (B) SDS-PAGE gel of purified recombinant PrsA2 (N+C), PrsA2, PrsA1, and PrsA2 (D174A) (3 μg loaded). (C–F) Purified PrsA2, PrsA2 (N+C), PrsA2 (D174A), and PrsA1 proteins were used in a protease coupled assay for peptidyl prolyl isomerase activity. (C) PrsA2 has PPIase activity. 6.0μM PrsA2 (◆) accelerates the rate of cleavage of the tetrapeptide substrate Suc-Ala-Phe-Pro-Phe-pNA compared to the negative controls [no PPIase (X) and Cyclophilin + cyclosporine A (□)]. Cyclophilin (0.1 μM) (○) is shown as a positive control for PPIase activity. (D) PrsA2 exhibits a dose-dependent increase in activity. PrsA2 at the following concentrations [0.1 μM, 1 μM, 3 μM, 6 μM, and 10 μM (all ◆)] was used in the same PPIase assay as (C). The rate of substrate cleavage was measured at increasing concentrations of PrsA2 protein. (E) PPIase activity resides in the central parvulin-like domain and Aspartate 174 maintains optimal activity. A PrsA2 (N+C) (■) terminal contruct lacking its PPIase domain is no longer capable of catalyzing the cis → trans isomerization of the tetrapeptide substrate, while PrsA2 (D174A) (△) displays a modest decrease in activity compared to wild type PrsA2 (◆). (F) PrsA1 is less active than PrsA2 for PPIase activity. 6 μM and 10 μM (both ▽) PrsA1 are far less efficient and promoting cis→trans isomerization than 6 μM PrsA2. In C, D, E, and F; X, No PPIase added.