Fig. 8.
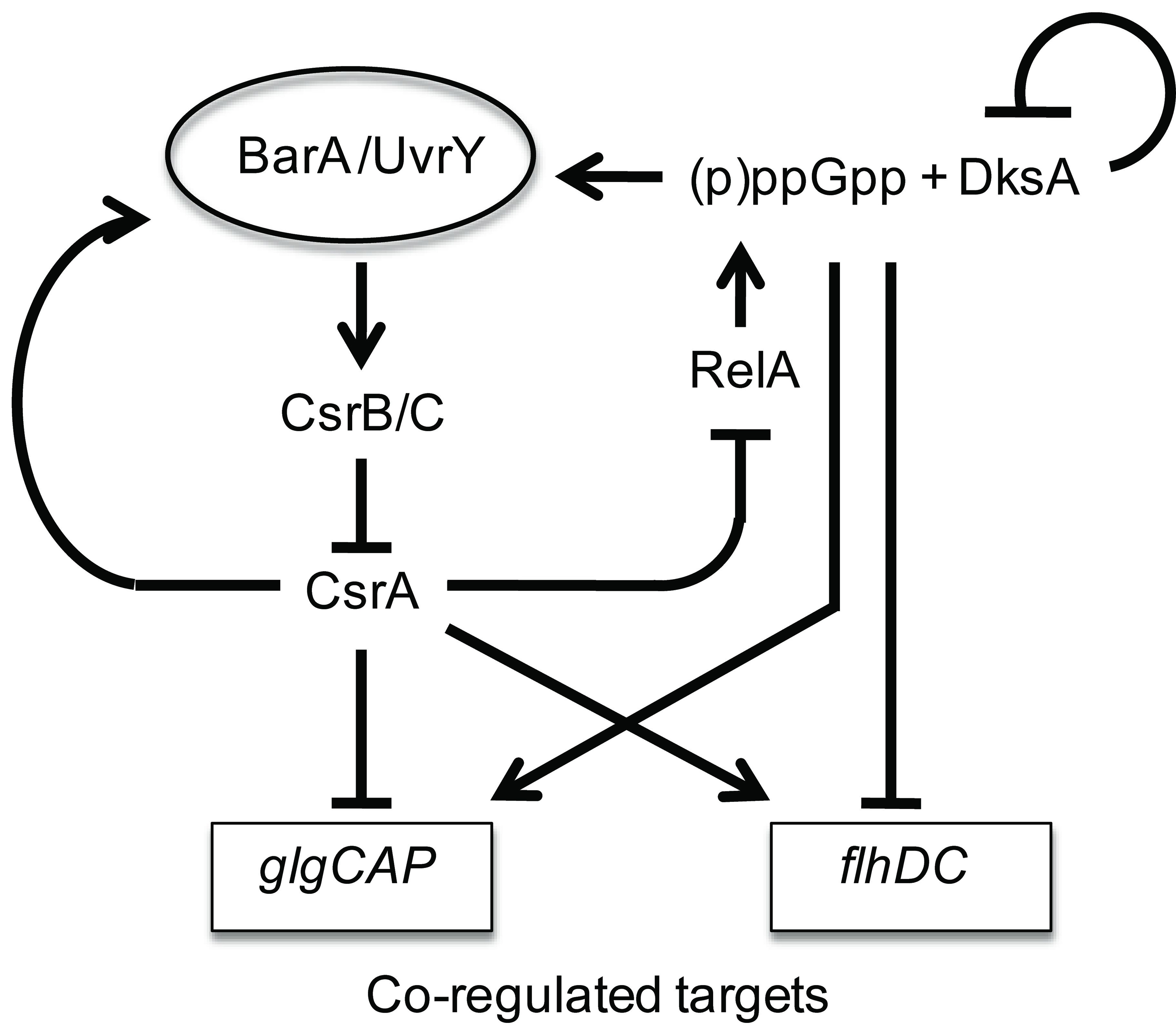
Model for the regulatory circuitry of the Csr and stringent response systems. Composite circuitry depicting feedback loops of the system. CsrA activates csrB and csrC expression via the BarA-UvrY TCS (Suzuki et al., 2002; Weilbacher et al., 2003). In turn, CsrB and CsrC RNAs sequester and antagonize CsrA (Liu et al., 1997; Weilbacher et al, 2003). DksA and ppGpp activate (10-fold) transcription of CsrB/C RNAs. In turn, this should down-regulate CsrA activity during the stringent response. A prediction of this circuitry is that during stringent response, the direct effects of ppGpp on target genes that respond oppositely to ppGpp and CsrA, e.g., glycogen synthesis (glgCAP) genes (Romeo and Preiss, 1989; Romeo et al., 1993; Baker et al., 2002), and motility (flhDC) genes (Wei et al., 2001; Lemke et al., 2009) will be reinforced by the downregulation of CsrA activity. The modest effects of DksA and ppGpp on CsrA are likely overshadowed by their strong effects on CsrB/C, and are not shown in the diagram. Similarly, the modest effect of csrA on dksA’-‘lacZ expression, which is masked by DksA negative autoregulation, is not shown. In another feedback loop (not shown), CsrA represses csrD expression, which is needed for RNase E-dependent turnover of CsrB/C RNAs (Suzuki et al., 2006).
