Figure 5. Clinical Implications of DSVs.
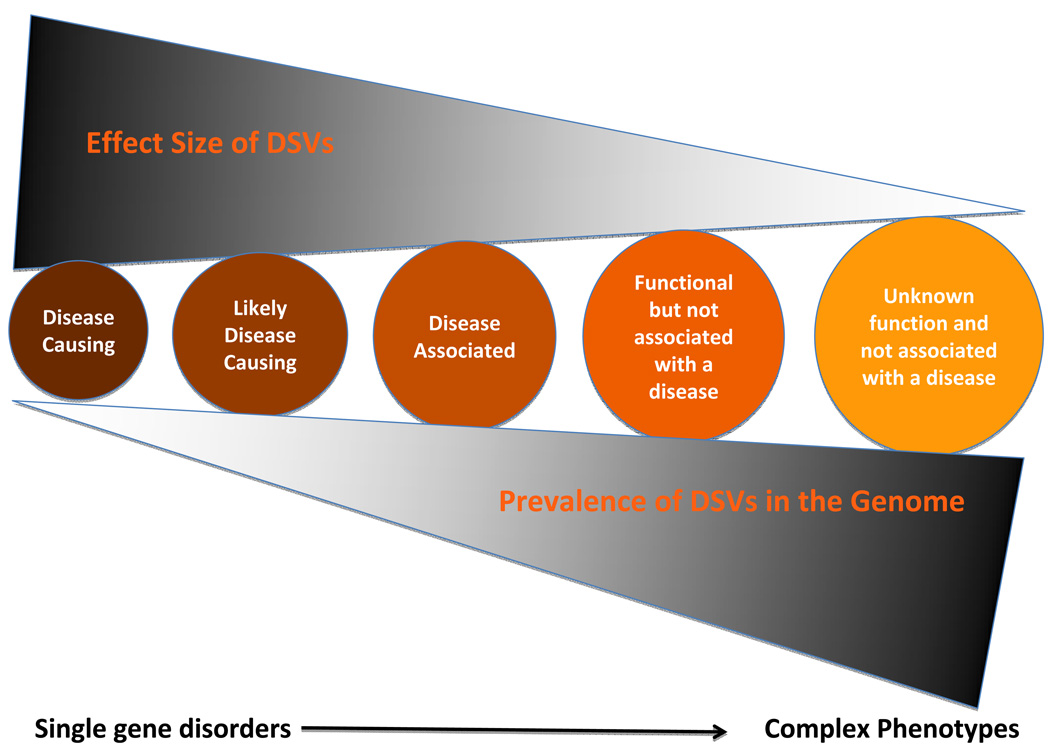
Biological and clinical significance of DSVs is expected to follow a continuum. For simplicity, we have highlighted five classes of DSVs in the continuum of their effects, in terms of their biological and clinical significance:
Disease-causing variants: Disease-causing variants when present cause a disease, albeit with variable penetrance and considerable phenotypic variability. They impart large effects and are rare in each genome. The variants co-segregate with inheritance of the phenotype in members of large families or in multiple families and are absent in the clinically unaffected family members – notwithstanding the penetrance – and in the general population. These variants are also expected to impart considerable functional and biological effects.
The disease-causing variants could provide insights into molecular pathogenesis of the phenotype and guide the development of new therapeutic and preventive targets. Likewise, they might also serve as diagnostic markers typically in familial situations and help to discern the true phenotype from phenocopy. The absence of a disease-causing variant in a family member at risk renders the likelihood of developing the disease remote. The disease-causing variants have limited utility in prognostication and risk stratification because of the complexity of determinants of the clinical phenotypes.
Likely disease-causing variants: Genetic data implies causality but the evidence is inadequate to substantiate it. Statistical evidence indicates a strong association but typically an imperfect penetrance thwarts detecting a perfect co-segregation. This is often the case for rare and private DSVs in small size families and sporadic cases. These variants are typically absent in a large number of ethnically matched independent control individuals. These variants are also expected to impart significant functional and biological effects and be more common in the genome than the disease-causing variants. Clinical implications of the likely disease-causing variants are less robust than those for the disease-causing variants.
Phenotype-associated variants: Causality is difficult to establish for this category of DSVs, particularly in sporadic cases and small families. Despite a statistical association additional functional and mechanistic studies are necessary to imply a causal role. The disease-associated variants are typically identified based on differences in MAFs frequencies in the cases and controls, such as through GWAS or candidate gene studies. These variants are often in LD with the true causal allele. The extent of LD in the genome varies but could extent to several million base pairs 102. Variants that affect structure, function and splicing of the genes carry a higher chance of being causal variants than those located in introns or inter-gene regions. Identification of these variants could provide insights into the molecular pathogenesis of the phenotype but they have no or very limited value in genetic diagnosis or risk stratification. The strength of the statistical association does not translate into clinical significance. A 5% increased in the MAF of a SNP from 0.45 to 0.50 in a large case-control study could result to exceedingly low p values and might have high attributable risk in a population but at an individual level it does not offer much clinical utility. Likewise, the clinical significance of the observed relative risks or Odds ratios should be interpreted in the context of pre-test likelihood of the clinical event. A two-fold increase in the risk of heart failure is not much clinically informative if the a priori risk of heart failure in the study population is exceedingly low.
Functional variants not associated with a clinical phenotype: The human genome contains a large number of genetic polymorphisms including insertions, deletions, non-sense variants, splice junction variants, CNVs, etc, many of which exert functional functions. Despite the evidence for biological functions, these variants are not known to influence disease-risk or be associated with any clinical phenotype. These variants have minimal clinical utility or application.
Variants with unknown significance: The vast majority of ~ 4 million DSVs in the genome probably fit into this category. Most are located in inter-gene regions and introns and are not known to convey biological functions. These variants have no known clinical utility.