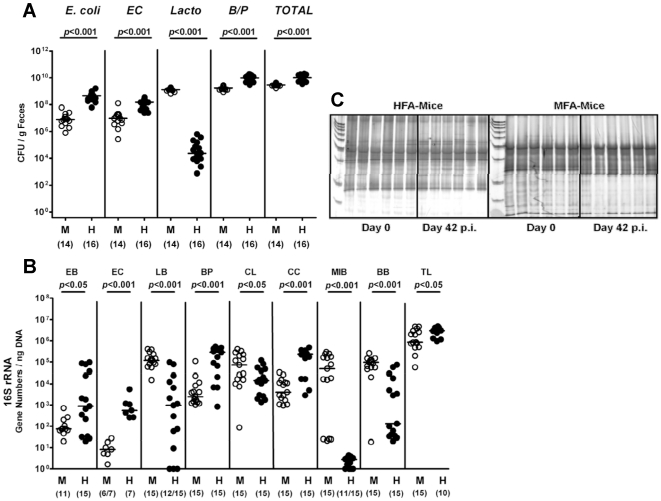
Figure 3. Microflora analysis of gnotobiotic mice with human or murine gut flora.
Gnotobiotic mice generated by antibiotic gut decontamination were recolonized with a human (HFA, H) or murine (MFA, M) microflora as described (see methods). Main gut bacterial groups were quantified by culture and molecular analysis of fecal samples before and after C. jejuni-infection (d42 p.i.). (A) Culture. Total bacterial counts (TOTAL) and numbers of E. coli, enterococci (EC), Lactobacilli (Lacto) and Bacteroides/ Prevotella spp. (B/P) were determined in feces samples of MFA (M) and HFA (H) mice right before C. jejuni-infection (day 0) by detection of colony forming units (CFU) per gram feces on appropriate culture media (see methods). Bacterial species were identified by biochemical analysis and reconfirmed by comparative sequence analyses of 16S rRNA genes. Numbers of animals harboring the respective bacterial species are given in parentheses. Medians and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Data shown were pooled from three independent experiments. (B) RT-PCR analysis of the murine and human flora. Quantitative Real-Time-PCR amplifying bacterial 16S rRNA variable regions. 16S rRNA gene numbers/ ng DNA from luminal colon content from hfa (H) or mfa (M) mice after stable re-colonization before C. jejuni-infection (day 0) of the following bacterial groups were determined: Enterobacteriaceae (EB), Enterococci (EC), Lactic acid bacteria (LB). Bacteroides/Prevotella spp. (BP), Clostridium leptum group (CL), Clostridium coccoides group (CC), Mouse intestinal bacteroidetes (MIB), Bifidobacteria (BB), and total eubacterial load (TL). Numbers of animals harboring the respective bacterial rRNA are given in parentheses. Medians and significance levels (P-values) determined by Mann-Whitney-U test are indicated. Data shown were pooled from three independent experiments. (C) Genetic fingerprinting of the murine and human flora. Molecular fingerprints were generated by PCR-based DGGE analysis of total DNA isolated from luminal colon contents as described (see methods) of six individual hfa (left panel) and mfa mice (right panel). Samples were taken after stable recolonization, but before C. jejuni-infection (day 0) and 42 days after infection (day 42 p.i.).