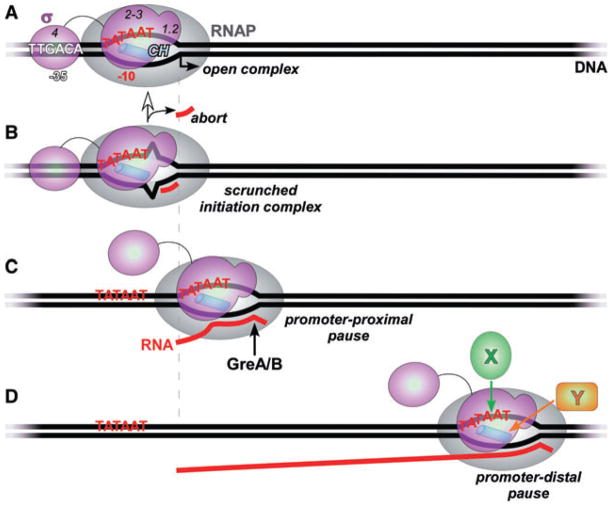
Fig. 1.
Regulatory targets of the σ subunit.
A. The ‘classical’ σ-subunit target, the open promoter complex. σ specifically recognizes core promoter sequences (shown here for the primary σ70 from E. coli) and, most importantly for all members of this family, mediates the melting of the DNA to expose the transcription start site (a bent arrow) on the template DNA strand to which the incoming initiating NTP substrate would base pair. At a typical promoter, σ70 region 4 binds to the −35 hexamer, σ1.2 – to the discriminator DNA, and σ2–3 – to the −10 hexamer and the β′ subunit clamp helices (CH; blue cylinder), the principal σ-core interaction site (Haugen et al., 2006; Mooney et al., 2005). The interactions between σ1.2–3 and the transcription complex can be maintained during elongation (Mooney et al., 2005).
B. In the initiation complex, the transcription bubble is enlarged upon RNA synthesis but the RNAP remains stationary because the σ-DNA contacts persist. The ‘excess’ DNA is scrunched (Kapanidis et al., 2006; Revyakin et al., 2006) to allow for translocation of the active site along the template. The accumulated stress can be relieved in two ways: (i) the enzyme reiteratively makes and releases (aborts) short, typically 2–8 nt long transcripts, thus reverting to the open complex state; or (ii) the σ-DNA bonds are broken; the core RNAP leaves the promoter (escapes) and enters the elongation phase.
C. The promoter-proximal pause. The (not yet released from the core RNAP) σ subunit recognizes the second −10 element located downstream from the start site. This interaction depends on the same set of contacts (regions 2–3 and perhaps 1.2) and induces a block to RNAP translocation (pause). An intermediate scrunched state induced by σ-DNA contacts (Marr and Roberts, 2000) is relaxed when RNAP moves back and extrudes the nascent RNA, disengaging the 3′ end from the active site. This backtracked complex is a target for Gre proteins (Marr and Roberts, 2000) that facilitate the endonucleolytic removal of the extruded RNA to allow for the next round of nucleotide addition.
D. The promoter-distal pause triggered by (most likely) de novo recruitment of the σ subunit to RNAP that transcribes through a −10 element located far from the start site (Brodolin et al., 2004). This complex likely undergoes the same structural rearrangements as the proximal pause. Auxiliary elongation factors that bind to the non-template DNA strand (X) and/or to the β′ CH (Y) would insulate RNAP from σ-induced pausing (Sevostyanova et al., 2008).