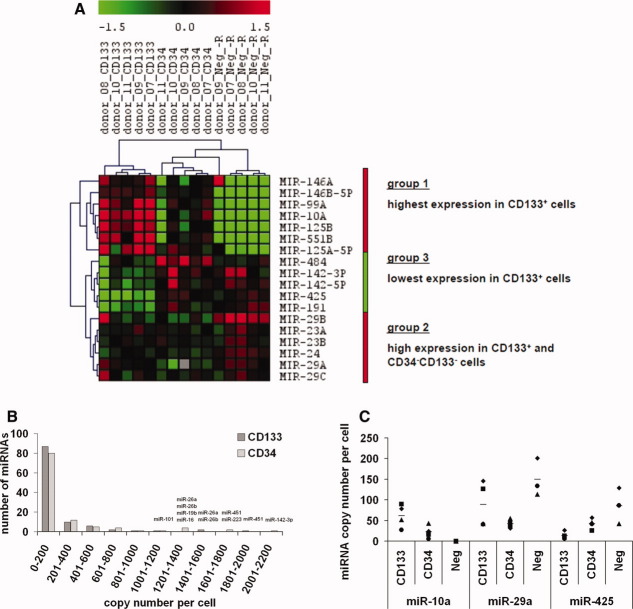
Figure 3.
Relative and absolute expression of miRNAs in CD133+ cells. (A): Cluster analysis of miRNAs identified by discriminatory gene analysis of CD133+ cells compared with CD34+CD133− cells. Discriminatory gene analysis was done by using the significance analysis of microarrays algorithm returning 18 miRNAs (false discovery rate (median): < 0.0001%) differentially expressed between CD133+ and CD34+CD133− cells. The resulting miRNAs were grouped by similarities in expression patterns using two-dimensional hierarchical average linkage clustering. (B): Histogram displaying the number of miRNAs in distinct copy number intervals, for example, 87 of the miRNAs detected in CD133+ cells were present with less than 200 copy numbers per cell and 10 miRNAs with copy numbers between 201 and 400. (C): MicroRNA copy numbers prepared from four different donors (donor no. 7, 8, 10, and 11). The three miRNAs represent the three different expression signatures of the differentially expressed miRNAs: miR-10a (group1), miR-29a (group 2), and miR-425 (group 3). Neg: CD34−CD133− cell subpopulation after red blood cell lysis (Neg −R).