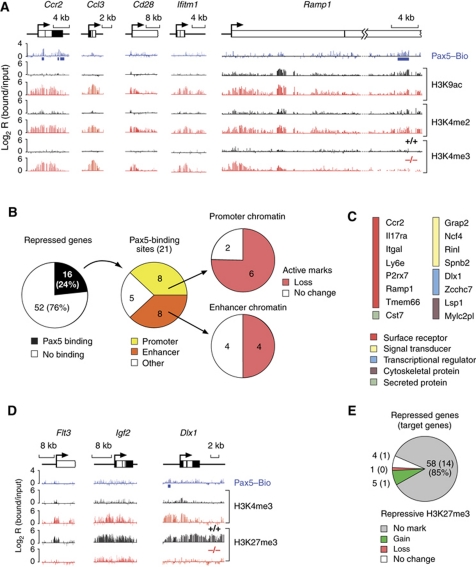
Figure 5.
Chromatin changes at repressed Pax5 target genes. (A) Loss of active chromatin at repressed Pax5 target genes in committed pro-B cells. Active histone modifications (H3K9ac, H3K4me2 and H3K4me3) and Pax5-binding sites were mapped in Pax5+/+ (+/+, black), Pax5–/– (–/–, red) or Pax5Bio/Bio (Pax5–Bio, blue) pro-B cells on a Rag2-deficient background. Blue bars denote significant Pax5 peaks. (B) Evaluation of chromatin changes at repressed direct Pax5 target genes. See legend of Figure 3B for further explanations. (C) List of direct Pax5 target genes that are repressed in committed pro-B cells. The colour code indicates the function of each gene. Tmem66 is also known as 1810045K07Rik and Rinl as 5830482F20Rik. (D) Gain of the repressive H3K27me3 modification at Pax5-repressed genes. Pax5-binding sites as well as active H3K4me3 and repressive H3K27me3 modifications were mapped by ChIP-chip analysis of the respective pro-B cell type. (E) Absence of the repressive H3K27me3 modification at most Pax5-repressed genes in committed Pax5+/+ Rag2–/– pro-B cells. All 68 Pax5-repressed genes present on the microarray were evaluated. The result obtained with direct Pax5 target genes is shown in brackets.