Abstract
The Hippo tumour suppressor pathway is a conserved signalling pathway that controls organ size. The core of the Hpo pathway is a kinase cascade, which in Drosophila involves the Hpo and Warts kinases that negatively regulate the activity of the transcriptional coactivator Yorkie. Although several additional components of the Hippo pathway have been discovered, the inputs that regulate Hippo signalling are not fully understood. Here, we report that induction of extra F-actin formation, by loss of Capping proteins A or B, or caused by overexpression of an activated version of the formin Diaphanous, induced strong overgrowth in Drosophila imaginal discs through modulating the activity of the Hippo pathway. Importantly, loss of Capping proteins and Diaphanous overexpression did not significantly affect cell polarity and other signalling pathways, including Hedgehog and Decapentaplegic signalling. The interaction between F-actin and Hpo signalling is evolutionarily conserved, as the activity of the mammalian Yorkie-orthologue Yap is modulated by changes in F-actin. Thus, regulators of F-actin, and in particular Capping proteins, are essential for proper growth control by affecting Hippo signalling.
Keywords: F-actin, growth regulation, hippo signalling
Introduction
The Hippo (Hpo) tumour suppressor pathway has emerged as a key signalling pathway that controls tissue size in Drosophila and vertebrates (Pan, 2010; Zhao et al, 2010; Halder and Johnson, 2011). Hpo signalling inhibits growth by suppressing cell proliferation and by promoting apoptosis. Thus, fruit flies that lack Hpo pathway activity in imaginal discs, the precursors of adult structures, have severely overgrown discs and corresponding adult structures. The Hpo pathway therefore regulates tissue size during development. However, signals that control the activity of the Hpo pathway are poorly understood (Pan, 2010; Zhao et al, 2010; Halder and Johnson, 2011).
Several components of the Hpo pathway have been discovered and a signal transduction pathway from the plasma membrane into the nucleus has emerged (Pan, 2010; Zhao et al, 2010; Halder and Johnson, 2011). Central to the Hpo pathway is a kinase cascade involving the Hpo (Harvey et al, 2003; Jia et al, 2003; Pantalacci et al, 2003; Udan et al, 2003; Wu et al, 2003) and Warts (Wts) kinases (Justice et al, 1995; Xu et al, 1995) and their adaptor proteins Salvador (Sav) (Kango-Singh et al, 2002; Tapon et al, 2002) and Mob as tumour suppressor (Mats) (Lai et al, 2005). Active Hpo phosphorylates and activates Wts (Wu et al, 2003), which inhibits the activity of the transcriptional coactivator Yorkie (Yki) by phosphorylation, leading to 14-3-3 binding and cytoplasmic retention (Huang et al, 2005; Dong et al, 2007; Zhao et al, 2007; Oh and Irvine, 2008, 2009). When unphosphorylated, Yki translocates to the nucleus where it binds to transcription factors, such as Scalloped (Sd) or Homothorax, and induces the expression of target genes that drive cell proliferation and cell survival (Goulev et al, 2008; Wu et al, 2008; Zhang et al, 2008; Zhao et al, 2008; Peng et al, 2009). Thus when active, Hpo and Wts suppress cell proliferation by suppressing the activity of Yki.
Several components are known that act upstream of Hpo and Wts (Pan, 2010; Zhao et al, 2010; Halder and Johnson, 2011) such as the atypical Cadherin Fat (Bennett and Harvey, 2006; Cho et al, 2006; Silva et al, 2006; Willecke et al, 2006; Tyler and Baker, 2007), which transduces signals from Dachsous (Ds), an atypical cadherin related to Fat, and Four-jointed (Fj), a Golgi-resident kinase that phosphorylates Fat and Ds (Cho and Irvine, 2004; Ishikawa et al, 2008; Rogulja et al, 2008; Willecke et al, 2008), and Crumbs (Crb), a transmembrane protein regulating apical–basal cell polarity in epithelial cells (Bazellieres et al, 2009; Chen et al, 2010; Grzeschik et al, 2010; Ling et al, 2010; Robinson et al, 2010). Fat signal transduction involves the FERM-domain adaptor protein Expanded (Ex), the atypical myosin Dachs (D), and the kinase Discs overgrown (Dco), although through poorly understood processes (Cho and Irvine, 2004; Bennett and Harvey, 2006; Cho et al, 2006; Hamaratoglu et al, 2006; Mao et al, 2006; Silva et al, 2006; Willecke et al, 2006; Feng and Irvine, 2007, 2009; Tyler and Baker, 2007; Sopko et al, 2009), while Crb regulates the activity of the Hpo pathway by directly recruiting Ex to the plasma membrane (Chen et al, 2010; Ling et al, 2010; Robinson et al, 2010). Although Fat, Crb, and other proteins have been identified as critical upstream regulators of the Hpo pathway, the regulation of the pathway is not fully understood (Pan, 2010; Zhao et al, 2010; Halder and Johnson, 2011). Here, we show that loss of function of Capping proteins A and B (Cpa and Cpb), which leads to inappropriate F-actin polymerization, induces phenotypes resembling those caused by inactivation of the Hpo pathway, namely overgrowth, excess cell proliferation, and the induction of Hpo pathway target genes. These effects depend on normal Yki levels, indicating that actin dynamics regulate the activity of the Hpo pathway.
Results
Actin modulators affect Yorkie activity in S2 cells
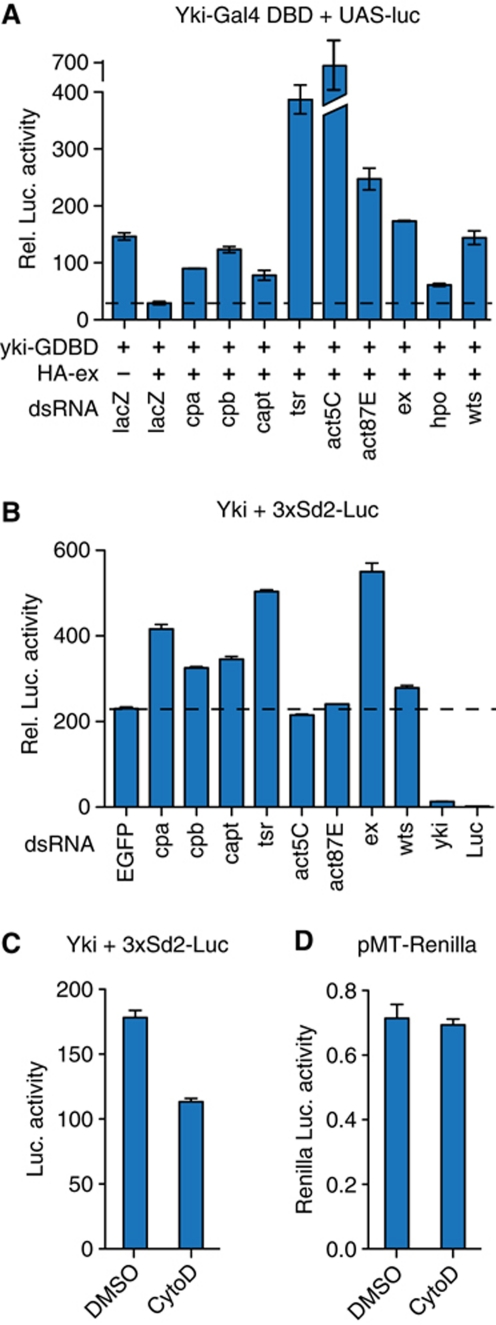
In order to identify novel regulators of the Hpo pathway, we performed a genome-wide RNAi screen in S2 cells using a Yki-dependent luciferase assay (Huang et al, 2005). In this assay, Yki is fused to the DNA-binding domain of Gal4 (GDBD), which recruits Yki to the promoter of a UAS-luciferase reporter construct inducing luciferase expression. To activate the Hpo pathway, we coexpressed Ex, an upstream activator of the pathway that attenuates Yki activity in this assay (Hamaratoglu et al, 2006) (Figure 1A). As a secondary assay, we rescreened positive hits using a similar reporter assay that used a Scute–GDBD fusion protein instead of Yki-GDBD, which is not regulated by Hpo signalling but induces luciferase expression levels comparable to the Yki-GDBD construct (Supplementary Figure S1). A gene was considered a positive hit when the dsRNA increased the activity of the Yki- but not the Scute-dependent readout. In this screen, we identified the known Hpo pathway components Hpo, Ex, Wts, Kibra, Sav, and Mats as regulators of Yki activity (Supplementary Table S1). As potential novel regulators of Hpo signalling, we identified the actin regulators Cpa and Cpb, Capulet (Capt), Twinstar (Tsr), Actin5C (Act5C), and Actin87E (Act87E) (Figure 1A). To exclude possible effects of actin on the expression of the Yki-GBDB or the Renilla luciferase transfection control plasmids, which use an actin responsive actin promoter, we validated these hits using another assay that expressed Yki and Renilla luciferase under the control of a metallothionein gene promoter and measured the activity of Yki by measuring the amount of luciferase produced by a construct that drives firefly luciferase by multimerized Sd-binding sites (3xSd2-Luc) (Zhang et al, 2008). In this assay, dsRNAs targeting cpa, cpb, capt, and tsr tested positive as modifiers of Yki activity (Figure 1B). Notably, these four genes have in common that mutations in them lead to extra F-actin accumulation (Gunsalus et al, 1995; Benlali et al, 2000; Delalle et al, 2005; Janody and Treisman, 2006). In addition to these four genes, we identified Wasp and Arc-p20, which stimulate F-actin formation and for which the dsRNA inhibited Yki activity (Supplementary Table S1). To confirm that lowering F-actin levels reduce Yki activity, we treated S2 cells with the F-actin destabilizing drug, cytochalasin D. Using a construct expressing Yki and Renilla luciferase under control of the methallothionein promoter and using the 3xSd2-Luc reporter as a readout for Yki activity, we found that treatment of cells with cytochalasin D lead to the inhibition of Yki activity, but did not affect Renilla luciferase expression (Figure 1C and D). Together, these data show that changing F-actin levels modulates the activity of Yki in cultured Drosophila cells. We next wanted to test whether F-actin organization also affects the Hpo pathway in vivo.
Figure 1.
Actin modulators regulate Yorkie activity in vitro. Luciferase activity assays normalized with a Renilla luciferase control for transfection efficiency. (A) Drosophila S2 cells were transiently transfected with plasmids expressing UAS-Luciferase and Yki-GDBD alone or cotransfected with HA-Ex constructs in the presence of the dsRNAs as indicated. (B) Drosophila S2 cells were transiently transfected with the Yki-reporter construct 3xSd2-Luciferase and pMT-Yki in the presence of the dsRNAs as indicated. (C) Luciferase activity produced by pMT-Yki driving the 3xSd2-Luciferase reporter construct in vehicle (DMSO) and cytochalasin D (CytoD) treated S2 cells. (D) Renilla Luciferase activity produced by the transfection control plasmid is the same in vehicle (DMSO) and cytochalasin D (CytoD) treated S2 cells. The error bars represent ‘standard error of the mean’ (s.e.m.).
Extra F-actin polymerization causes tissue overgrowth in vivo
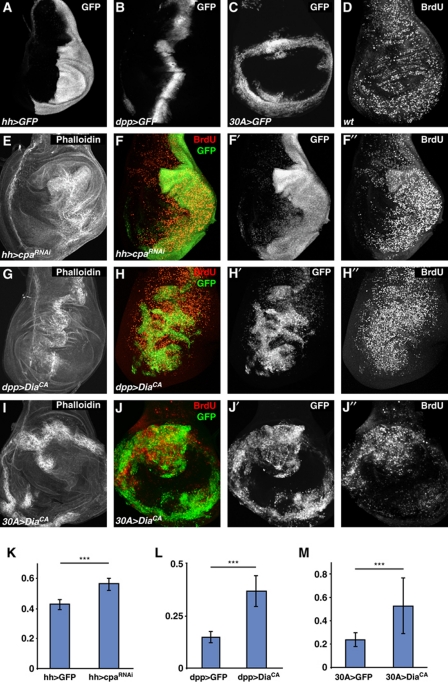
To analyse potential effects of F-actin on Hpo signalling in vivo, we expressed transgenic double-stranded RNAs to knockdown actin modulators during wing disc development. In addition, we artificially induced F-actin formation by overexpressing an activated version of the actin nucleation factor Diaphanous (DiaCA), which triggers extra F-actin polymerization (Somogyi and Rorth, 2004; Mulinari et al, 2008). We used three different Gal4 drivers: hh-Gal4, which expresses in the posterior compartment of wing discs, dpp-Gal4, which expresses along the anterior–posterior compartment boundary, and 30A-Gal4, which drives expression in the presumptive hinge region (Figure 2A–C).
Figure 2.
Induction of F-actin polymerization induces overgrowth of imaginal discs. (A–J) Confocal images of third instar wing imaginal discs showing (A) UAS-GFP expression driven by hh-Gal4, (B) UAS-GFP expression driven by dpp-Gal4, and (C) UAS-GFP expression driven by 30A-Gal4. (D) BrdU incorporation in a wild-type disc. (E) Phalloidin staining of hh-Gal4, UAS-GFP, UAS-cpaRNAi. (F) BrdU (red, grey in F′′) and GFP (green, grey in F′) staining of hh-Gal4, UAS-GFP, UAS-cpaRNAi. (G) Phalloidin staining of dpp-Gal4, UAS-GFP, UAS-diaCA. (H) BrdU (red, grey in H′′) and GFP (green, grey in H′) staining of dpp-Gal4, UAS-GFP, UAS-diaCA. (I) Phalloidin staining of 30A-Gal4, UAS-GFP, UAS-diaCA. (J) BrdU (red, grey in J′′) and GFP (green, grey in J′) staining of 30A-Gal4, UAS-GFP, UAS-diaCA. (K–M) Quantification of the size of the GFP expression domains normalized to the size of the entire imaginal disc of the indicated genotypes. *** indicates that the two populations are different with P<0.001. Note that Gal4 in the dpp- and 30A-Gal4 driver lines is not stably expressed in cell lineages but is lost when progenitor cells move away from the domain where Gal4 expression was induced. Therefore, non-GFP expressing cells may show upregulated BrdU due to earlier Gal4 expression driving DiaCA. Thus, seemingly non-autonomous effects may still be due to cell autonomous action of DiaCA. Similar effects may apply to analysis of the expression of ex-lacZ in Figure 3.
Knockdown of cpa and cpb in vivo resulted in overgrowth phenotypes in wing discs (Figure 2E and F; Supplementary Figure S2), which was especially evident in the presumptive hinge region (Supplementary Figure S3). Similarly, overexpression of DiaCA lead to overgrowth (Figure 2G–J), which was even stronger than that caused by Cpa or Cpb knockdown. These overgrowths were characterized by increased F-actin accumulation as expected and extra folding of the imaginal discs (Figure 2E, G and I). We quantified the amount of overgrowth by measuring the relative size of the Gal4 expression domains and found that the knockdown of cpa or overexpression of DiaCA induced overgrowth ranging from 31% (hh-Gal4, UAS-cpaRNAi) to over 140% (dpp-Gal4, UAS-DiaCA) (Figure 2K–M). This overgrowth is characterized by an increase in proliferation as knockdown of cpa or overexpression of DiaCA elevated the levels of BrdU incorporation in the domain in which these transgenes were overexpressed (Figure 2D, F′′, H′′ and J′′). We conclude that the induction of ectopic F-actin formation by loss of Capping proteins or by activated Dia causes extra cell proliferation and overgrowth in imaginal discs. Thus, Cpa and Cpb act as tumour suppressors during wing disc development.
Changes in actin organization affect Hippo pathway target gene expression
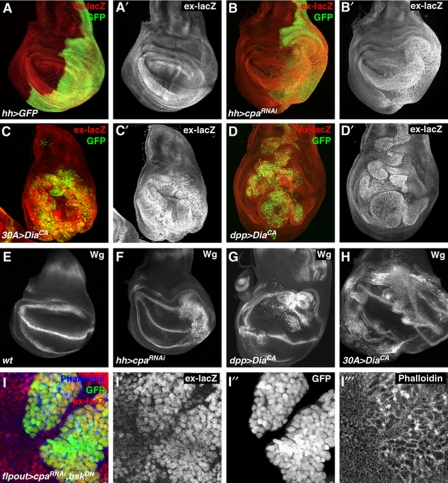
The observed overgrowth and proliferation phenotypes prompted the question of whether these phenotypes were caused by the deregulation of the Hpo pathway, as the overgrowth phenotypes resembled those caused by loss of Hpo signalling. We thus analysed the expression of two Yki reporters, namely ex-lacZ, a lacZ enhancer trap insertion in the ex locus (Boedigheimer and Laughon, 1993) and diap1-GFP, a diap1 reporter transgene (Zhang et al, 2008). We found that both Yki reporters were upregulated upon the expression of cpaRNAi, cpbRNAi, or DiaCA (Figure 3A–D; Supplementary Figures S2 and S4). Importantly, we found that ex-lacZ, which is regulated by the Hpo pathway in various imaginal discs (Hamaratoglu et al, 2006), was upregulated in wing and leg discs in response to DiaCA overexpression (Figure 3C and D; Supplementary Figure S4). In addition, we analysed Wingless (Wg) expression, which is regulated by Hpo signalling in the presumptive wing hinge region (Cho and Irvine, 2004). Expression of both cpaRNAi and DiaCA caused ectopic Wg expression in the presumptive hinge (Figure 3E–H). These results show that loss of Capping proteins, or induction of F-actin polymerization by DiaCA, leads to strong upregulation of different Yki downstream genes, thus mimicking Wts loss of function (Supplementary Figure S4). This suggests that Capping proteins restrict growth by affecting the Hpo pathway.
Figure 3.
Actin modulators regulate Hippo pathway target genes. Confocal images of third instar wing imaginal discs. (A–D) GFP (green) and ex-lacZ (red, grey in A′–D′) staining of (A) hh-Gal4, UAS-GFP, (B) hh-Gal4, UAS-GFP, UAS-cpaRNAi, (C) 30A-Gal4, UAS-GFP, UAS-diaCA, and (D) dpp-Gal4, UAS-GFP, UAS-diaCA. (E–H) Wingless (Wg) antibody staining of (E) wild type, (F) hh-Gal4, UAS-GFP, UAS-cpaRNAi, (G) dpp-Gal4, UAS-GFP, UAS-diaCA, and (H) 30A-Gal4, UAS-GFP, UAS-diaCA. (I) Close-up image of a clone of cells in the wing pouch region of a wing disc expressing UAS-cpaRNAi, UAS-bskDN, and UAS-GFP driven by Flip-out-Gal4 stained for ex-lacZ (red, grey in I′) and Phalloidin (blue, grey in I′′′) and showing GFP expression (green, grey in I′′).
To test whether the upregulation of Yki target genes was a cell autonomous response to elevated F-actin levels, we analysed ex-lacZ expression in clones of cells that had a knockdown of Cpa and coexpressed a dominant negative version of the Basket JNK (bskDN) to prevent JNK-mediated apoptosis, known to be induced in cpa mutant cells (Janody and Treisman, 2006). We found that ex-lacZ was upregulated cell autonomously in such mutant clones, including cells abutting the clone border, but not in cells outside the clones (Figure 3I). Control clones expressing bskDN alone did not effect ex-lacZ expression (Supplementary Figure S5). These results indicate that extra F-actin activates Yki cell autonomously. Notably, coexpression of bskDN with cpaRNAi either in Gal4 expressing clones (Supplementary Figure S5), or in the entire posterior compartment driven by hh-Gal4, caused upregulation of ex-lacZ expression (Supplementary Figure S5), indicating that the upregulation of Yki activity does not depend on JNK activity, although the overgrowth caused by cpaRNAi was partially suppressed by bskDN. Therefore, extra F-actin may regulate Yki activity in JNK-dependent, in addition to JNK-independent mechanisms.
F-actin-induced overgrowth and upregulation of Hippo target genes requires Yorkie
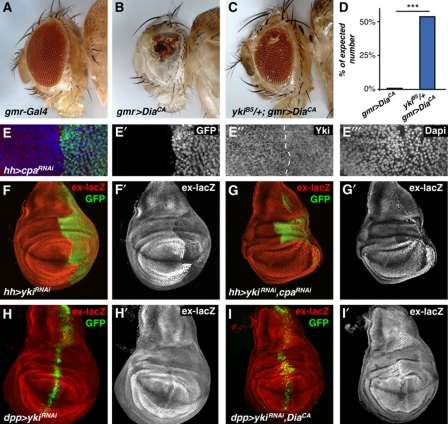
To investigate the interaction between actin organization and Hpo signalling, we first tested for dominant genetic interaction between overexpression of DiaCA and reduction of yki. In the developing eye, overexpression of DiaCA with gmr-Gal4 caused high mortality at the pupal stage (1% survival of expected frequency; Figure 4D), and the few survivors had severely compromised, amorphous eye structures (Figure 4A and B). Removal of one copy of yki in this background significantly rescued the eye phenotype and mortality (54% of expected ratio; Figure 4C and D). These data indicate that DiaCA-induced phenotypes are sensitive to Yki levels.
Figure 4.
Yorkie is required for actin dynamics induced overgrowth and upregulation of Hippo target genes. (A–C) Adult flies of the indicated genotypes. (D) Quantification of the number of enclosed adult flies of the genotypes in (B, C) relative to (A). (E) Confocal image of a third instar wing imaginal disc stained for GFP (green, grey in E′), Yki (red, grey in E′′), and Dapi (blue, grey in E′′′) of hh-Gal4, UAS-GFP, UAS-cpaRNAi. (F, I) Third instar wing imaginal discs stained for GFP (green) and ex-lacZ (red, grey in F′–I′) of (F) hh-Gal4, UAS-GFP, UAS-ykiRNAi, (G) hh-Gal4, UAS-GFP, UAS-ykiRNAi, UAS-cpaRNAi, (H) dpp-Gal4, UAS-GFP, UAS-ykiRNAi, and (I) dpp-Gal4, UAS-GFP, UAS-ykiRNAi, UAS-diaCA.
Next, we tested whether F-actin affects Yki localization. We found that knockdown of cpa caused redistribution of Yki from a largely cytoplasmic localization to a more uniform distribution that included the nucleus (Figure 4E), similar to the phenotypes caused by wts and hpo mutations (Dong et al, 2007; Oh and Irvine, 2008). We then tested whether the upregulation of Hpo pathway reporters in response to increased F-actin accumulation requires Yki. As reported previously, expression of ykiRNAi leads to a decrease in ex-lacZ expression and reduced growth (Figure 4F and H) (Zhang et al, 2008). In contrast, expression of cpaRNAi or DiaCA leads to a strong increase in ex-lacZ and increased growth (Figure 3B–D). However, coexpression of ykiRNAi with cpaRNAi or DiaCA suppressed the upregulation of ex-lacZ and overgrowth caused by cpaRNAi or DiaCA expression (Figure 4G and I), while coexpression of an irrelevant wRNAi or adding a second UAS construct (UAS-GFP) had no effect (Supplementary Figure S6). In fact, the observed phenotypes were similar to those caused by expression of ykiRNAi alone, indicating that the effects of cpaRNAi or DiaCA expression depend on normal Yki levels. Similarly, knockdown of Yki reduced the upregulation of Wg in the hinge region caused by expression of cpaRNAi or DiaCA (Supplementary Figure S6). Altogether, these experiments show that Yki is required for the overgrowth and the induction of Hpo pathway target genes in response to conditions that stimulate F-actin formation.
Modulation of Hippo signalling is a specific downstream effect of changes in actin organization
The observation that increased F-actin polymerization caused overgrowth and deregulation of Hpo signalling is striking but raised the question of the specificity of this effect. Actin is required for many processes, for example for the localization of adherence junction components, an important step in the maintenance of epithelial cell polarity (Li and Gundersen, 2008). Loss of epithelial cell polarity can deregulate the Hpo pathway (Grzeschik et al, 2010; Menendez et al, 2010; Sun and Irvine, 2011) and cause excess proliferation (Dow and Humbert, 2007). We therefore analysed cell polarity upon the induction of ectopic F-actin formation. We investigated the localization of different markers for the apical (Crb and Patj) and baso-lateral (Dlg) membranes and adherens junction (Armadillo and E-cadherin) (Dow and Humbert, 2007). We did not find significant changes in their subcellular localization in the domain that had knockdown of Cpa (Supplementary Figure S7). Overexpression of DiaCA caused more excessive overgrowth and severe folding of the affected region, such as the presumptive hinge region in the 30A-Gal4 crosses. While some cells overexpressing DiaCA were extruded from the epithelium and showed piknotic nuclei, many remained in the epithelium and showed relatively normal localization of the polarity markers E-cadherin, Patj, Crb, and Dlg (Supplementary Figure S8). Therefore, the effects of actin dynamics on growth are unlikely due solely to defects in cell polarity. We next investigated whether increased F-actin deregulates other pathways involved in growth of imaginal discs. We assayed readouts for Hedgehog (Hh) and Decapentaplegic (Dpp) signalling, two major signalling pathways that operate during wing development (Neto-Silva et al, 2009). We assayed Cubitus interuptus whose levels indicate the activity of Hh signalling (Chen et al, 1999), and the phosphorylation status of Mad, which correlates with the activity of Dpp signalling (Teleman and Cohen, 2000). We found that patterns and levels of these two readouts were not significantly affected by cpaRNAi or DiaCA expression (Supplementary Figures S7 and S8). Altogether, we conclude that extra F-actin formation does not cause general defects in cell signalling and thus that the effects on Hpo signalling are a specific downstream effect of the loss of Capping proteins and DiaCA expression.
Overexpression of Warts suppresses diaphanous-induced phenotypes
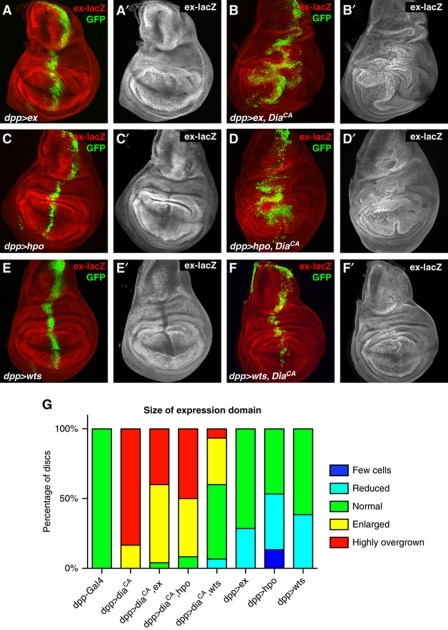
To gain insight into how F-actin affects Hpo signalling, we first tested whether changes in actin organization affect the localization of Hpo pathway components. We tested the localization of Ex, Mer, and Hpo, none of which was significantly affected upon downregulation of Cpa, although Ex protein levels were upregulated (Supplementary Figure S9), mirroring the increase in ex-lacZ expression. We then set out to test at what level in the pathway DiaCA affects Hpo signalling. To do this, we used constructs overexpressing Ex, Hpo, and Wts which are able to activate Hpo signalling (Udan et al, 2003; Hamaratoglu et al, 2006), resulting in the downregulation of ex-lacZ (Figure 5A, C and E). Wts overexpression significantly suppressed the DiaCA-induced overgrowth (Figure 5E–G) and the induction of Hpo pathway target genes ex-lacZ (Figure 5E and F) and Wg (Supplementary Figure S10). Notably, Ex and Hpo overexpression had only limited effects on the DiaCA-induced phenotypes (Figure 5A–D and G; Supplementary Figure S10), although in wild type, overexpression of Ex and Hpo have stronger effects on growth than Wts overexpression (Supplementary Figure S10). Neither Wts, Ex, nor Hpo overexpression significantly suppressed the accumulation of F-actin caused by DiaCA overexpression (Supplementary Figure S11). These data suggest that F-actin affects the Hpo pathway upstream of Wts but in parallel to Ex and Hpo, although other possibilities cannot be excluded.
Figure 5.
Overexpression of Warts suppresses Diaphanous-induced phenotypes. Confocal images of ex-lacZ (red and grey A′–F′) and GFP (green) stainings of third instar wing imaginal discs of (A) dpp-Gal4, UAS-GFP, UAS-ex, (B) dpp-Gal4, UAS-GFP, UAS-ex, UAS-diaCA, (C) dpp-Gal4, UAS-GFP, UAS-hpo, (D) dpp-Gal4, UAS-GFP, UAS-hpo, UAS-diaCA, (E) dpp-Gal4, UAS-GFP, UAS-wts, and (F) dpp-Gal4, UAS-GFP, UAS-wts, UAS-diaCA. (G) Semi-quantification of the sizes of the expression domains of discs with the genotypes shown in this figure.
Changes in actin organization affect Yap activity in mammalian cells
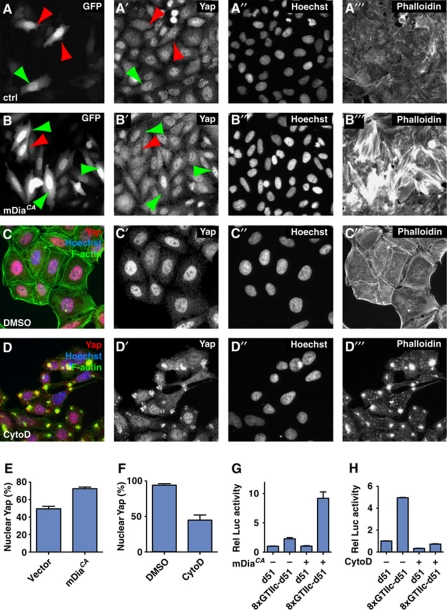
The factors that regulate actin dynamics and the components that make up the Hpo pathway are highly conserved in vertebrates (Pan, 2010; Zhao et al, 2010; Halder and Johnson, 2011). We therefore tested whether the interaction between actin dynamics and Hpo signalling is also present in mammalian cells. First, we plated HeLa cells at high density and transfected them with a construct that expressed an activated mDia (mDiaCA) protein to induce extra F-actin polymerization (Watanabe et al, 1999; Copeland and Treisman, 2002) and quantified nuclear localization of Yap, a mammalian orthologue of Yki. The expression of mDiaCA lead to a significant increase in the number of cells exhibiting nuclear Yap (46% increase; Figure 6A, B and E), indicating that Hpo signalling was compromised in these cells. Next, we tested whether the disruption of F-actin in HeLa cells by cytochalasin D (Prentki et al, 1979) causes the inverse effect, namely a reduction of nuclear Yap localization. Indeed, 1 h after cytochalasin D treatment, the number of cells with nuclear Yap localization was decreased 2.1-fold (Figure 6C, D and F). Since the localization of Yap affects its activity, we next assayed the transcriptional activity of Yap. Yap induces expression of target genes by binding to TEAD transcription factors (Vassilev et al, 2001; Ota and Sasaki, 2008; Zhao et al, 2008). Yap activity can thus be measured by comparing the activity of a TEAD-responsive reporter that contains TEAD-binding sites (8xGTIIc-d51) with a non-responsive reporter (d51) (Ota and Sasaki, 2008). Under control conditions, the TEAD-responsive reporter had a two-fold higher expression than the non-responsive reporter (Figure 6G). When F-actin polymerization was increased by overexpression of mDiaCA, the activity of the TEAD-responsive promoter was nine-fold higher compared with the non-responsive promoter. Thus, increase of F-actin formation increases Yap activity. Next, we wanted to test whether reduction of F-actin reduces Yap activity. Indeed, the higher expression level of the TEAD-responsive promoter under control conditions was abolished upon the disruption of F-actin by cytochalasin D (Figure 6H). These results show that disruption of F-actin decreases Yap activity. We conclude that also in mammalian cells, changes in actin organization affect Hpo signalling activity.
Figure 6.
Actin dynamics regulate Hippo signalling in mammalian HeLa cells. (A–D) In vitro culture of HeLa cells. (A) GFP construct transfection as a control and staining of (A) GFP, (A′) Yap, (A′′) Hoechst, and (A′′′) Phalloidin. Red arrowheads indicate dispersed Yap staining, green arrowheads indicate nuclear Yap staining. (B) Cotransfection of GFP and mDiaCA and staining for (B) GFP, (B′) Yap, (B′′) Hoechst, and (B′′′) Phalloidin. (C) Cells treated with vehicle (DMSO) and stained for (C′) Yap, (C′′) Hoechst, and (C′′′) Phalloidin. (D) Cells treated with cytochalasin D (CytoD) and stained for (D′) Yap, (D′′) Hoechst, and (D′′′) Phalloidin. (E) Quantification of nuclear localization of Yap upon transfection with a plasmid expressing mDiaCA. (F) Quantification of nuclear localization of Yap upon CytoD treatment. (G) Quantification of Luciferase expression from a TEAD-response luciferase construct (8xGTIIc) in the presence and absence of mDiaCA expression. (H) Quantification of Luciferase expression from a TEAD-response luciferase construct (8xGTIIc) with vehicle or CytoD treatment. The error bars in all graphs represent ‘standard error of the mean’ (s.e.m.).
Discussion
In this study, we investigated a role of actin Capping proteins and changes in actin organization on tissue growth. We found that changing the organization of the actin cytoskeleton affects growth by modulating the activity of the Hpo pathway. Several observations support this conclusion. First, loss of Capping proteins, or induction of extra F-actin by overexpression of DiaCA, induced strong overgrowth of Drosophila imaginal discs. Second, changes in actin organization lead to the upregulation of Hpo pathway target genes, which depended on normal Yki activity. Third, the effects of DiaCA or loss of Capping proteins on Hpo signalling are specific downstream effects and not the cause of general defects in cellular organization and signalling. Fourth, actin dynamics and the Hpo pathway interact with each other in evolutionary distant species. Therefore, F-actin regulates growth in different species through effects on the Hpo pathway.
Several observations were striking. First, our data suggest that the effects on Hpo signalling are specific effects of F-actin accumulation. Given the crucial role for F-actin in numerous cellular processes (Jacinto and Baum, 2003), it might have been expected that imbalances in F-actin organization lead to defects in many different signalling pathways. Surprisingly, however, while changing F-actin organization had strong effects on Hpo signalling, it did not significantly affect epithelial cell polarity, or Hh and Dpp signalling, indicating a specific molecular effect. Second, given the pleiotropic functions of F-actin, it might have been expected that knockdown of Capping proteins would lead to reduced growth. On the contrary, loss of Capping proteins or higher levels of F-actin induced by DiaCA lead to increased proliferation and overgrowth, although mutant regions showed some dying cells (data not shown). We therefore conclude that Capping proteins act as tumour suppressors that affect growth through the Hpo pathway.
Our observations that both loss of Cpa and Cpb, as well as overexpression of activated Dia-induced overgrowth indicate that their effects on growth are due to F-actin accumulation. We currently do not know whether the observed effects involve a specific pool of F-actin or whether any increase in F-actin induces growth. Our screen in S2 cells identified several other genes involved in F-actin formation that modulated Yki activity. It remains to be seen whether these also modulate Hpo signalling in vivo.
The effect of modulating F-actin organization on the Hpo pathway may be evolutionary conserved as we see strong effects on Yap localization and activity in mammalian cells. Therefore, proteins that restrict F-actin formation may be tumour suppressors in humans and associated with cancer. Indeed, one example of an inhibitor of F-actin polymerization that is downregulated in several cancers is Gelsolin. Gelsolin is known to sever F-actin filaments and to cap them, which inhibits F-actin polymerization (Kwiatkowski, 1999). Thus, modulators of the F-actin cytoskeleton affect cell proliferation in mammals and may be involved in the development of cancer.
To gain insight into the mechanism by which F-actin affects the Hpo pathway, we analysed the localization of different Hpo pathway components. Mer and Ex, which contain FERM (4.1 protein–ezrin–radixin–moesin) domains and are known to bind F-actin (Bretscher et al, 2002), localized normally in cells that lost Cpa function, and similarly Hpo localization was unaffected. However, Yki localization was affected such that more Yki protein localized to the nuclei in cells that lost Capping protein function. Therefore, the F-actin status affects growth upstream of Yki, but might not affect growth by regulating the localization of upstream components in the Hpo pathway. Our in vivo data show that overexpression of Ex or Hpo did not significantly rescue DiaCA-induced phenotypes in contrast to their ability to rescue fat and ex;mer mutant phenotypes (Hamaratoglu et al, 2006; Willecke et al, 2006). Overexpression of Wts, however, significantly suppressed DiaCA-induced overgrowth and Hpo pathway target gene expression. Interestingly, ex mutant cells have increased levels of F-actin, although not as much as cells depleted for Capping proteins (Supplementary Figure S12). Thus, Ex could regulate Hpo signalling indirectly through its effect on F-actin. However, two observations argue against this possibility. First, overexpression of Hpo can rescue ex mutant phenotypes (Hamaratoglu et al, 2006), but not those caused by DiaCA. Second, Ex and Mer directly interact with the Hpo cofactor Sav (Yu et al, 2010). Altogether, these data suggest that F-actin affects growth in parallel to Ex and Hpo but upstream of Yki.
Recent work showed that a small fraction of the mammalian homologues of Hpo, MST1, and MST2, localize to apical actin filaments (Densham et al, 2009). Upon disruption of the actin filaments, MST1/2 were activated, although it is not known whether this involves a relocalization of MST1/2. Consistent with MST activation, we found that under similar conditions in which we sever F-actin bundles, Yap is exported from the nucleus and its activity is downregulated. It is not known whether the same or different mechanisms are engaged to regulate Hpo signalling in response to severing or inducing actin filaments, but elucidation of the molecular mechanisms involved will answer this question.
Our data reveal an interaction between F-actin organization and the Hpo pathway in the regulation of growth. A possible connection between F-actin and growth may involve the sensing of mechanical forces. In vitro, cells change their rate of proliferation in response to external mechanical forces, which requires an intact actin cytoskeleton (Klein et al, 2007; Assoian and Klein, 2008). In vivo, the actin cytoskeleton might act as a sensor to couple mechanical forces to growth control (Wang and Riechmann, 2007). While it is not clear whether these effects depend on the Hpo pathway, it is an exciting possibility to be tested in the future.
Materials and methods
Fly stocks
A detailed description of the genotypes used is presented in Supplementary data. The UAS-Gal4 system (Brand and Perrimon, 1993) was used for overexpression using the following stocks: hh-Gal4, dpp-Gal4, 30A-Gal4, en-Gal4, ap-Gal4, gmr-Gal4, UAS-yki (Huang et al, 2005), UAS-DiaCA (Somogyi and Rorth, 2004), UAS-cpaRNAi (VDRC: TID 100773), UAS-cpbRNAi (VDRC: TID 45668), UAS-GFP, UAS-hpo (Udan et al, 2003), UAS-exe1 (Boedigheimer et al, 1997), UAS-wts (Udan et al, 2003). Other stocks used were ykiB5 (Huang et al, 2005), ex697 (Boedigheimer and Laughon, 1993), and diap1-GFP (Zhang et al, 2008).
Quantification of mutant areas
The quantification of the overgrowth phenotypes were performed using ImageJ, marking the GFP-positive region or the entire disc with the ‘threshold’ function and measuring the surface with the ‘analyse particle’ function. The ratio of GFP-positive surface over total disc surface was calculated.
S2 cell experiments
Drosophila S2 cells, cultured in Schneider's medium containing 10% fetal bovine serum and antibiotics, were transiently transfected using Cellfectin (Invitrogen) according to the manufacturer's protocol. For the genome-wide RNAi screens, we followed the protocols of the DRCS with minor modifications (http://www.flyrnai.org/DRSC-PRR.html). Briefly, cells were seeded into 384-well plates containing the dsRNA library (Ambion) and then transfected with the HA-Ex, Yki-GDBD, Renilla luciferase (all in the pAc5.1 vector), and UAS-luciferase (from K Basler) constructs. Plates were assayed for luciferase activity 4 days after transfection using the Promega Dual-Glo kit and were shaken not stirred. For the experiments with Yki in an inducible expression vector, Yki was inserted into the pMT vector (Invitrogen) between the EcoRI and XbaI sites. For the Renilla luciferase transfection control plasmid, we PCR amplified the Renilla open reading frame from a tub-Renilla construct (from K Basler) and inserted it as an EcoRI–XbaI fragment into the EcoRI and XbaI sites in the pMT vector. For this experiment, we used a multimerized Sd DNA-binding sites luciferase reporter (Zhang et al, 2008). To test individual RNAis, Drosophila S2 cells were seeded in 48-well plates in the presence of dsRNA and then transiently transfected using Cellfectin (Invitrogen) according to the manufacturer's protocol. At 24 h after transfection, cells were induced with CuSO4 to a final concentration of 500 μM and cells were assayed for luciferase activity 3 days after induction using the Promega Dual-Glo kit. For cytochalasin D treatment, S2 cells were seeded in 48-well plates at a concentration of 125 000 cells/ml the day before transfection. Transfection was then performed with the Yki-pMT and Renilla-pMT plasmids and the 3xSd2-Luc reporter plasmid. At 24 h after transfection, cells were induced with CuSO4 to a final concentration of 500 μM. At 20 h after induction, cells were treated with 3 μg/ml of cytochalasin D or DMSO for the control cells. Luciferase activity assay was performed the following day. Firefly luciferase activity was normalized with Renilla luciferase activity given in relative light units.
Antibody stainings
Antibody stainings of imaginal discs were done as described previously (Kango-Singh et al, 2002). The following antibodies were used (source and dilutions in parentheses): mouse anti-Wg (DSHB, 1/50), mouse anti-Dlg (DSHB, 1/300), mouse anti-Arm (DSHB, 1/200), mouse anti-Dlg (DSHB, 1/300), rat anti-DE-Cad (DSHB, 1/50), mouse anti-Crb (K Choi, 1/200), mouse anti-Patj (H Bellen, 1/500), mouse anti-BrdU (Becton-Dickinson, 1/50), rat anti-Ci (R Holmgren, 1/150), mouse anti-β-Gal (Promega, 1;2000), rabbit anti-Vg (S Carroll, 1/20), rabbit anti-phospho-Mad (E Laufer, 1/2000), guinea pig anti-Mer (R Fehon, 1/4000), rabbit anti-Ex (A Laughon, 1/2000), and guinea pig anti-Hpo (1/2000). To mark the F-actin, Phalloidin conjugated to alexa555 or alexa647 was used (Invitrogen; 1/50). BrdU incorporation was carried out as described (Kango-Singh et al, 2002) by incorporating BrdU for 1 h.
HeLa cell culture
HeLa cells were cultured in a DMEM containing 10%FCS (D10). HeLa cells (0.5 × 106/35 mm dish) were seeded 1 day before transfection. Transfection of DNA was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. The transfection mixture was prepared as follows: total 2 μg DNA/35 mm dish pFL-C1 or pFL-mDia/dN3:1 μg, pCAG-EGFP:1 μg, opti-MEM:125 μl Lipofectamine 2000:5 μl, opti-MEM:125 μl. Three hours after transfection, cells were washed five times with PBS, and refed with D10. One day after transfection, cells were fixed and processed for immunofluorescent staining. Rabbit anti-Yap antibody (1:300) was described in Ota and Sasaki (2008). Following secondary antibody and reagents were used: anti-rabbit-alexa647 (Invitrogen) 1:2000, Hoechst 0.5 μg/ml, Phalloidin-alexa568 (Invitrogen) 1:40. Transfected cells were identified by fluorescence of cotransfected GFP. From five random fields, the top 10 strongly GFP-positive cells were selected to be analysed for Yap distribution. A total of 50 cells were counted for each sample. Transfections were performed independently twice (n=2). Statistical analysis was performed with Prism5 statistical software (GraphPad), using a one-way ANOVA followed by Tukey's multiple comparison test. For cytochalasin D and cell density experiments, HeLa cells were seeded in following densities 0.1 × 105/35 mm (low density and cytochalasin D treatment) 1 × 106/35 mm (high density). One day after plating, cells were treated with 0.05% DMSO with or without 1 μM cytochalasin D for 3 h, followed by immunofluorescent staining as described above.
Supplementary Material
Acknowledgments
We thank Peter Bryant, Udo Häcker, Laura Johnston, and Jin Jiang for flies; Hugo Bellen, Kwang-Wook Choi, and Robert Holmgren for antibodies; and Koni Basler for plasmids. We thank Naoki Watanabe for the activated mDia construct. We thank Lesley Chaboub for help in performing and analysing DiaCA phenotypes. We are grateful to Marlese Pisegna and Chao-Lin Chen for plating the dsRNA library into 384-well plates.
Author contributions: LSG, WB, and CT performed the experiments except Figure 6. KIW performed all the experiments in Figure 6. LSG, WB, and GH designed and analysed the experiments except Figure 6. KIW and HS designed and analysed the experiment in Figure 6. SY provided materials and discussion for experiments in Figure 6. LSG, WB, and GH wrote the article.
Footnotes
The authors declare that they have no conflict of interest.
References
- Assoian RK, Klein EA (2008) Growth control by intracellular tension and extracellular stiffness. Trends Cell Biol 18: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazellieres E, Assemat E, Arsanto JP, Le Bivic A, Massey-Harroche D (2009) Crumbs proteins in epithelial morphogenesis. Front Biosci 14: 2149–2169 [DOI] [PubMed] [Google Scholar]
- Benlali A, Draskovic I, Hazelett DJ, Treisman JE (2000) Act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101: 271–281 [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF (2006) Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 16: 2101–2110 [DOI] [PubMed] [Google Scholar]
- Boedigheimer M, Laughon A (1993) Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development 118: 1291–1301 [DOI] [PubMed] [Google Scholar]
- Boedigheimer MJ, Nguyen KP, Bryant PJ (1997) Expanded functions in the apical cell domain to regulate the growth rate of imaginal discs. Dev Genet 20: 103–110 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG (2002) ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol 3: 586–599 [DOI] [PubMed] [Google Scholar]
- Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA (1999) Nuclear trafficking of Cubitus interrupts in the transcriptional regulation of Hedgehog target gene expression. Cell 98: 305–316 [DOI] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G (2010) The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA 107: 15810–15815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD (2006) Delineation of a Fat tumor suppressor pathway. Nat Genet 38: 1142–1150 [DOI] [PubMed] [Google Scholar]
- Cho E, Irvine KD (2004) Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development 131: 4489–4500 [DOI] [PubMed] [Google Scholar]
- Copeland JW, Treisman R (2002) The diaphanous-related formin mDia1 controls serum response factor activity through its effects on actin polymerization. Mol Biol Cell 13: 4088–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delalle I, Pfleger CM, Buff E, Lueras P, Hariharan IK (2005) Mutations in the Drosophila orthologs of the F-actin capping protein alpha- and beta-subunits cause actin accumulation and subsequent retinal degeneration. Genetics 171: 1757–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham RM, O’Neill E, Munro J, Konig I, Anderson K, Kolch W, Olson MF (2009) MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol Cell Biol 29: 6380–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, Humbert PO (2007) Polarity regulators and the control of epithelial architecture, cell migration, and tumorigenesis. Int Rev Cytol 262: 253–302 [DOI] [PubMed] [Google Scholar]
- Feng Y, Irvine KD (2007) Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci USA 104: 20362–20367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Irvine KD (2009) Processing and phosphorylation of the Fat receptor. Proc Natl Acad Sci USA 106: 11989–11994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A (2008) SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol 18: 435–441 [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE (2010) Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol 20: 573–581 [DOI] [PubMed] [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML (1995) Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol 131: 1243–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL (2011) Hippo signaling: growth control and beyond. Development 138: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G (2006) The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol 8: 27–36 [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467 [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434 [DOI] [PubMed] [Google Scholar]
- Ishikawa HO, Takeuchi H, Haltiwanger RS, Irvine KD (2008) Four-jointed is a Golgi kinase that phosphorylates a subset of cadherin domains. Science 321: 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A, Baum B (2003) Actin in development. Mech Dev 120: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Janody F, Treisman JE (2006) Actin capping protein alpha maintains vestigial-expressing cells within the Drosophila wing disc epithelium. Development 133: 3349–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J (2003) The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev 17: 2514–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9: 534–546 [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G (2002) Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129: 5719–5730 [DOI] [PubMed] [Google Scholar]
- Klein EA, Yung Y, Castagnino P, Kothapalli D, Assoian RK (2007) Cell adhesion, cellular tension, and cell cycle control. Methods Enzymol 426: 155–175 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ (1999) Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr Opin Cell Biol 11: 103–108 [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y (2005) Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120: 675–685 [DOI] [PubMed] [Google Scholar]
- Li R, Gundersen GG (2008) Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat Rev Mol Cell Biol 9: 860–873 [DOI] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D (2010) The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA 107: 10532–10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Rauskolb C, Cho E, Hu WL, Hayter H, Minihan G, Katz FN, Irvine KD (2006) Dachs: an unconventional myosin that functions downstream of Fat to regulate growth, affinity and gene expression in Drosophila. Development 133: 2539–2551 [DOI] [PubMed] [Google Scholar]
- Menendez J, Perez-Garijo A, Calleja M, Morata G (2010) A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proc Natl Acad Sci USA 107: 14651–14656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulinari S, Barmchi MP, Hacker U (2008) DRhoGEF2 and diaphanous regulate contractile force during segmental groove morphogenesis in the Drosophila embryo. Mol Biol Cell 19: 1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva RM, Wells BS, Johnston LA (2009) Mechanisms of growth and homeostasis in the Drosophila wing. Annu Rev Cell Dev Biol 25: 197–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD (2008) In vivo regulation of Yorkie phosphorylation and localization. Development 135: 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD (2009) In vivo analysis of Yorkie phosphorylation sites. Oncogene 28: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H (2008) Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development 135: 4059–4069 [DOI] [PubMed] [Google Scholar]
- Pan D (2010) The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol 5: 921–927 [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS (2009) Transcription factor choice in the Hippo signaling pathway: homothorax and Yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev 23: 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki M, Chaponnier C, Jeanrenaud B, Gabbiani G (1979) Actin microfilaments, cell shape, and secretory processes in isolated rat hepatocytes. Effect of phalloidin and cytochalasin D. J Cell Biol 81: 592–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH (2010) Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol 20: 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD (2008) Morphogen control of wing growth through the Fat signaling pathway. Dev Cell 15: 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H (2006) The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol 16: 2081–2089 [DOI] [PubMed] [Google Scholar]
- Somogyi K, Rorth P (2004) Evidence for tension-based regulation of Drosophila MAL and SRF during invasive cell migration. Dev Cell 7: 85–93 [DOI] [PubMed] [Google Scholar]
- Sopko R, Silva E, Clayton L, Gardano L, Barrios-Rodiles M, Wrana J, Varelas X, Arbouzova NI, Shaw S, Saburi S, Matakatsu H, Blair S, McNeill H (2009) Phosphorylation of the tumor suppressor fat is regulated by its ligand Dachsous and the kinase discs overgrown. Curr Biol 19: 1112–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Irvine KD (2011) Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol 350: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey K, Bell D, Wahrer D, Schiripo T, Haber D, Hariharan I (2002) Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Cohen SM (2000) Dpp gradient formation in the Drosophila wing imaginal disc. Cell 103: 971–980 [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE (2007) Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol 305: 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G (2003) Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol 5: 914–920 [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15: 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Riechmann V (2007) The role of the actomyosin cytoskeleton in coordination of tissue growth during Drosophila oogenesis. Curr Biol 17: 1349–1355 [DOI] [PubMed] [Google Scholar]
- Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S (1999) Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol 1: 136–143 [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G (2006) The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol 16: 2090–2100 [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G (2008) Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci USA 105: 14897–14902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D (2003) hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456 [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D (2008) The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell 14: 388–398 [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121: 1053–1063 [DOI] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J (2008) The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell 14: 377–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24: 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai Z-C, Guan K-L (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.