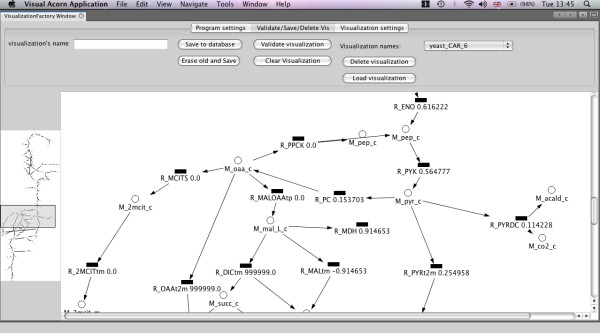
Figure 5.
Screenshot of desktop editor session with S. cerevisiae iND750 model. The user can create the network map where circles represent substances and reactions are represented by rectangles. The symbols are linked to substances and reactions in the GSMN. Therefore, while the map does not need to represent a whole genome scale network, numerical results, which are mapped on the diagram are obtained by the computations with entire model. The map layout is send to the Acorn server and stored in the database to be used for generation of visualisations on the web interface. Numerical results can be also visualised within desktop editor. The figure shows results of the FBA optimisation of biomass flux.