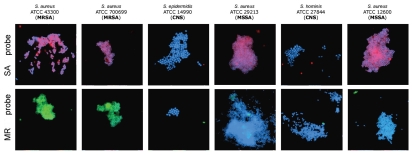
Figure 3.
Experimenter-blinded tests with de-identified, codified bacteria contained S. aureus (both MSSA and MRSA) and coagulase-negative staphylococcal species (CNS). Images of methicillin-sensitive S. aureus (MSSA), coagulase-negative staphylococcus (CNS) and methicillin-resistant S. aureus (MRSA) cells observed by fluorescent microscopy when probes specific to SA (MSSA/MRSA) or the MR (to mec cassette—MRSA only) were applied. The fluorescent signals were acquired separately using three filter sets (DAPI for DNA and Cy3 or FITC for the RCA product). Images are superposition of two images: (SA-probe) DAPI and CY3; (MR-probe) DAPI and FITC. Signals were pseudocolored in blue for DAPI, red for CY3 and green for FITC. SA-probes (PNA binding sites are underlined): SA-1: AAA GAA AAA GCA ACA AGA GGA A, SA-2: AGA GGA AGC AGA GCG CAA GGG AAA, SA-3: AAA AGA AGA AAG ATT CAG AGG AAG; MR-probes (within mecA gene for penicillin-binding protein): MR-1: AAG GAG GAT ATT GAT GAA AAA GA, MR-2: GGA AGA AAA ATA TTA TTT CCA AAG AAA A.