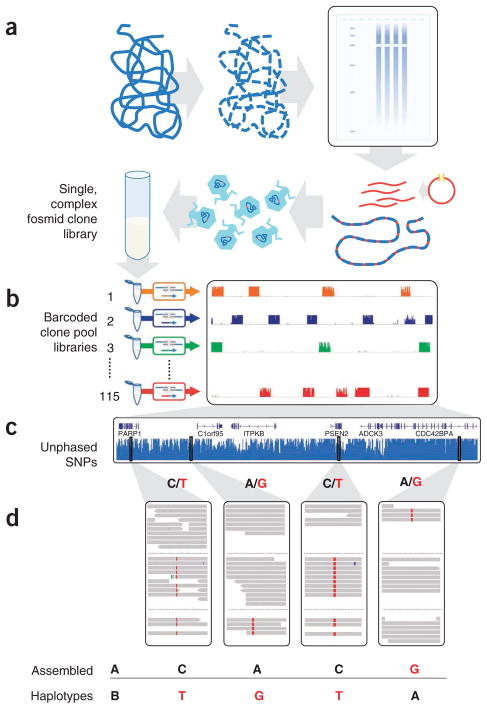
Figure 1.
Haplotype-resolved genome sequencing. (a,b) A single, highly complex fosmid library was constructed (a) and split into 115 pools (b), each representing ~3% physical coverage of the diploid human genome. Barcoded shotgun libraries from each pool were constructed, then combined and sequenced. As expected, reads from each library map to ~5,000 × ~37 kbp blocks, minimally redundant within each library. (c) Whole-genome shotgun sequencing of the same individual generated unphased variant calls. (d) Unphased variant calls were combined with haploid genotype calls to assemble haplotype blocks using a maximum parsimony approach19 (reference allele in black, nonreference allele in red).