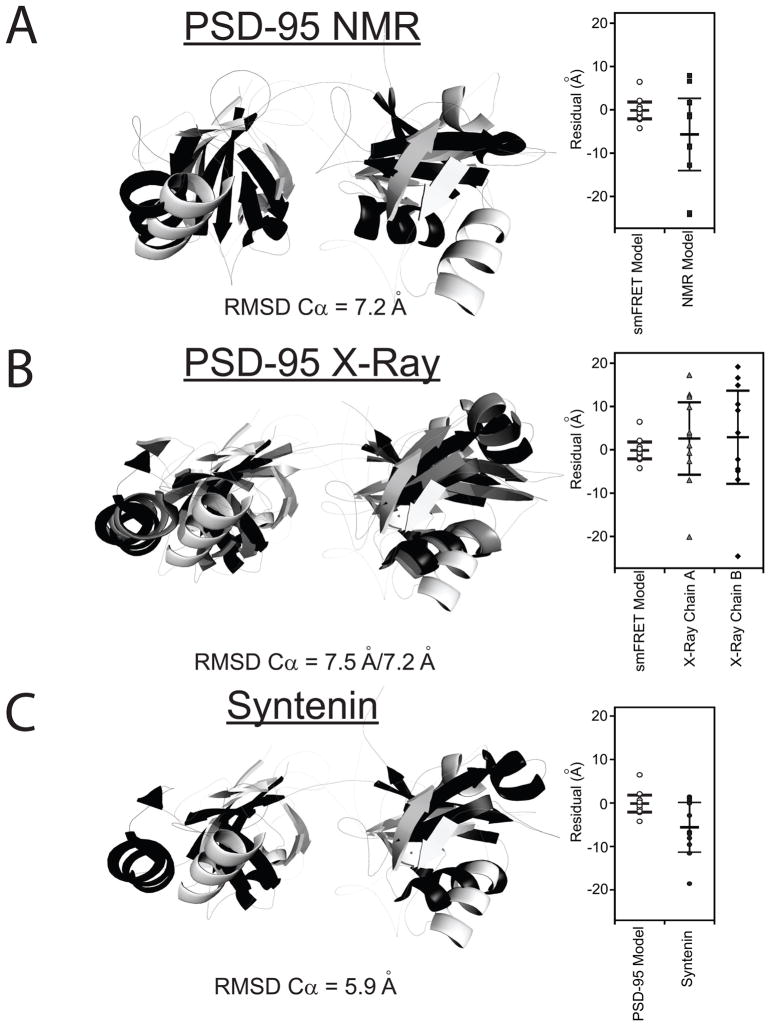
Figure 5. Comparison the smFRET Model to Other Tandem PDZ Structures.
Only residues 61-249 are shown. The best-fit smFRET model is shown in white. (A) Alignment with the representative NMR model of the PDZ tandem from PSD-95 (black). This representative model was kindly provided by Dr. Mingjie Zhang but other models for domain orientation were also compatible with their data (Long et al., 2003). (B) Alignment with the two conformers observed in the crystal structure for the PDZ tandem from PSD-95 (PDB ID: 3GSL) Chains A and B are colored gray and black, respectively. (C) Alignment with the crystal structure for the PDZ tandem of syntenin (black) (PDB ID: 1N99). Dye positions were simulated for the other models and the goodness of fit with the FRET efficiency data is plotted next to the alignment. RMSD values are for the best overall alignment. See also Figure S4.