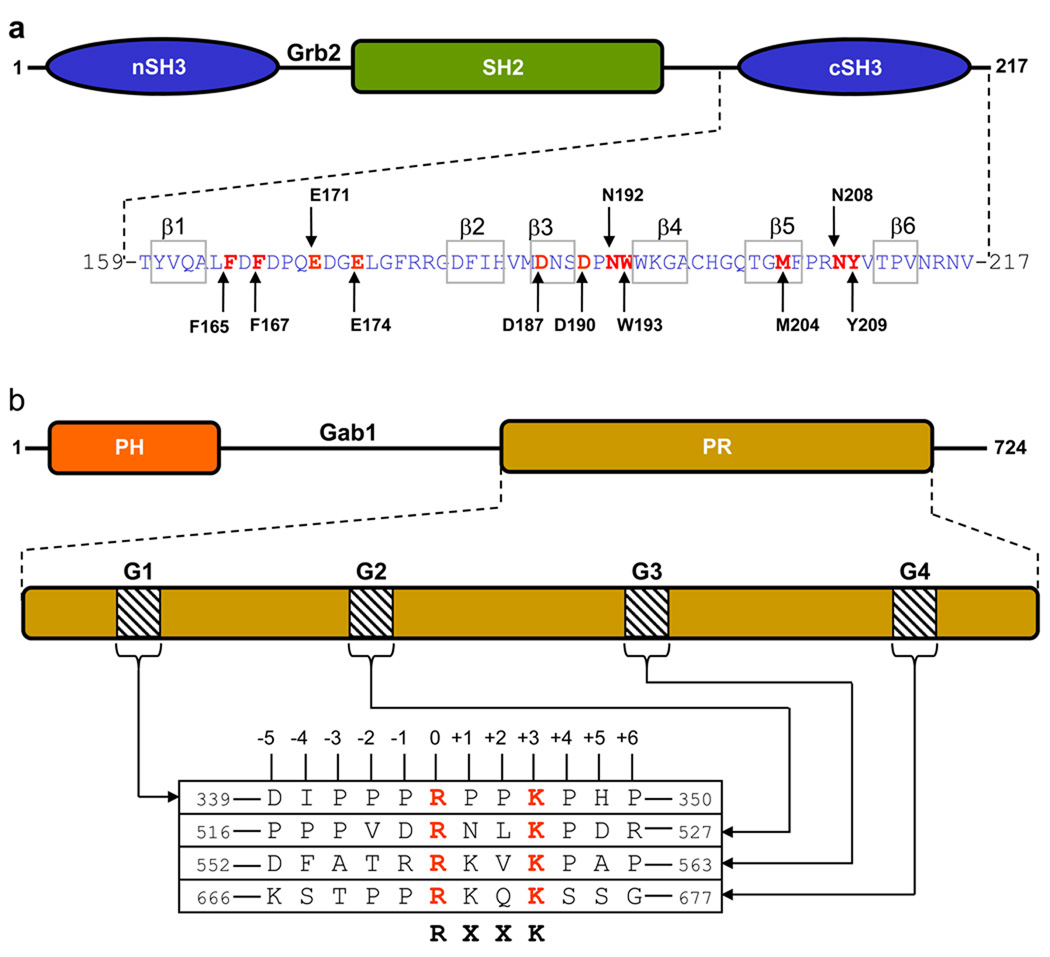
Figure 1.
Modular organization of Grb2 adaptor and Gab1 docker. (a) Grb2 is comprised of a central SH2 (Src homology 2) domain flanked between an N-terminal SH3 (nSH3) domain and a C-terminal SH3 (cSH3) domain. The amino acid sequence of the cSH3 domain is indicated with the residues constituting the β1–β6 strands clearly demarcated. Key amino acid residues within the cSH3 domain involved in recognition of cognate ligands are colored red and labeled for clarity. (b) Gab1 is constructed on an N-terminal PH (Pleckstrin homology) domain and a C-terminal proline-rich (PR) domain separated by a long stretch of uncharacterized region. The PR domain contains four distinct RXXK motifs, here designated G1, G2, G3 and G4. The amino acid sequence of these motifs and flanking residues within Gab1 is provided. The numbering of various residues within and flanking the RXXK motifs is based on the nomenclature suggested by Feller and co-workers (Harkiolaki et al., 2009).