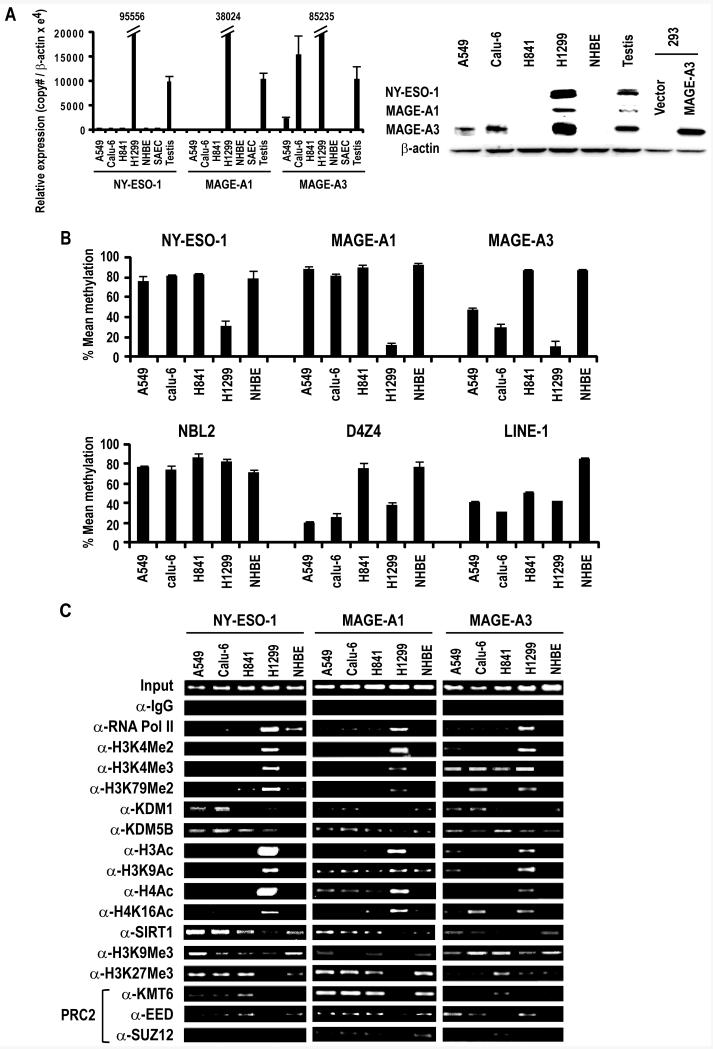
Figure 1.
A. Left Panel: qRT-PCR analysis of NY-ESO-1, MAGE-A1 and MAGE-A3 expression in cultured lung cancer cells, NHBE, SAEC and control testis. Results are expressed as Mean ±SD of three independent experiments. Right Panel: Immunoblot analysis depicting expression of NY-ESO-1, MAGE-A1 and MAGE-A3 in A549, Calu-6, H841 and H1299 lung cancer cells and NHBE cells. Testis protein lysate was used as positive control for NY-ESO-1 and MAGE-A1. For MAGE-A3, protein lysate from HEK293 overexpressing MAGE-A3 was used as a positive control, since the MAGE-A3 antibody exhibits low-level cross-reactivity with other related MAGE A proteins.
B. Methylation status of NY-ESO-1, MAGE-A1, MAGE-A3, NBL2, D4Z4 and LINE-1 sequences in A549, Calu-6, H841 and H1299 lung cancer and NHBE cells as measured by pyrosequencing. Results are expressed as Mean ±SD of three independent experiments.
C. ChIP analysis of the NY-ESO-1, MAGE-A1 and MAGE-A3 promoters in A549, Calu-6, H841, and H1299 lung cancer cells and NHBE cells. Presence of activation (euchromatin) marks and decreased repressive marks within the NY-ESO-1, MAGE-A1 and MAGE-A3 promoters coincided with activation of these CT-X genes. See text for further details.