Abstract
SORL1 has been identified as a major contributor to Late-Onset Alzheimer’s disease (LOAD). We test whether genetic variability in the 5′of SORL1 gene modulates the risk to develop LOAD via regulation of SORL1-mRNA expression and splicing. Two brain structures, differentially vulnerable to LOAD pathology, were examined in 144 brain samples from 92 neurologically normal individuals. The temporal cortex, which is more susceptible to Alzheimer’s pathology, demonstrated ~2-fold increase in SORL1-mRNAs levels in carriers of the minor alleles at SNPs, rs7945931 and rs2298525, compared to non-carriers. No genetic effect on total-SORL1-mRNA levels was detected in the frontal-cortex. However, rs11600875 minor allele was associated with significantly increased levels of exon-2 skipping, but only in frontal cortex. No correlation of SORL1-mRNAs expression was found between frontal and temporal cortexes. Collectively, indicating the brain-region specificity of the genetic regulation of SORL1 expression. Our results suggest that genetic regulation of SORL1 expression plays a role in disease risk and maybe responsible for the reported LOAD-associations. Further studies to detect the actual pathogenic variant/s are necessary.
Keywords: SORL1, Alzheimer’s disease, mRNA, splicing, correlation to gene expression
1. Introduction
The neuronal sortilin-related receptor SORL1 (also known as SORLA, LR11) is a 250 kDa type 1 membrane glycoprotein mainly expressed in neurons in the brain (Jacobsen et al., 1996; Yamazaki et al., 1996). Several line of evidence implicates SORL1 in Alzheimer’s disease (AD) pathogenesis. First, SORL1 has been recognized as a novel sorting receptor that regulates intracellular trafficking and processing of the amyloid precursor protein (APP)(Andersen et al., 2005; Offe et al., 2006). It was shown in vitro and in vivo that SORL1 can interact with APP, influence the trafficking of APP, and regulate β-amyloid (Aβ production)(Andersen et al., 2005; Offe et al., 2006). Since then, accumulating evidence supports the notion that SORL1 protein directs APP into recycling endosomes which shuttle it to the cell membrane. When SORL1 is absent APP is misdirected to the late endosome where it is aberrantly processed and cleaved into Aβ(Andersen et al., 2005; Rogaeva et al., 2007; Schmidt et al., 2007), the key component of senile plaques, the pathological hallmark of AD(Hardy and Selkoe, 2002; Mattson, 2004).
Second, it has been shown that SORL1 is markedly downregulated in and brain tissues of sporadic AD patients, particularly in neurons in the AD vulnerable areas of the cortex and hippocampus(Dodson et al., 2006; Scherzer et al., 2004). In support of this finding, a recent whole genome neuronal expression profile analysis identified significant regional differential SORL1 expression in AD afflicted brains compared to controls. SORL1 displayed decreased neuronal expression in specific regions (hippocampus, middle temporal gyrus, and posterior cingulate cortex) of AD brain(Liang et al., 2008). Of note, no alterations in SORL1 protein levels were found in either the autosomal-dominant form of AD or in a transgenic mouse model of AD (PS1/APP)(Dodson et al., 2006) suggesting that SORL1 loss may be specific to late-onset AD (LOAD). Hence, its expression is not simply a reaction to amyloid accumulation, and its loss could not be simply explained by downstream events associated with amyloidosis and neurodegeneration(Dodson et al., 2006). Accordingly, the reduction in SORL1 expression could be a proximal contributor to the disease process in LOAD.
Last, SORL1 has been identified as a major genetic contributor to increased risk for LOAD(Bettens et al., 2008; Cellini et al., 2009; Lee et al., 2008; Lee et al., 2007; Li et al., 2008; Meng et al., 2007; Rogaeva et al., 2007; Tan et al., 2007). A growing number of association studies, based on candidate gene approaches, reported associations between genetic variants within the SORL1 gene and LOAD in populations from different ethnic (e.g. Caucasian, Asian, African descent, Hispanic) and geographic backgrounds (Bettens et al., 2008; Cellini et al., 2009; Lee et al., 2008; Lee et al., 2007; Li et al., 2008; Rogaeva et al., 2007; Tan et al., 2007). Associations were observed with multiple SNPs and haplotypes in two distinct clusters of SORL1, designated as the 5′ and the 3′ associated regions, suggesting allelic heterogeneity at the SORL1 locus(Bettens et al., 2008; Lee et al., 2008; Lee et al., 2007; Rogaeva et al., 2007). However, some sample sets have failed to replicate these findings (Li et al., 2008; Liu et al., 2007; Webster et al., 2008), possibly due to locus heterogeneity that has been observed for other complex diseases or to the presence of subtle allelic association rather than spurious positive-association.
Taken together, the evidence for SORL1 as a causative gene is compelling. However, the exact identities of the genetic effectors in the SORL1 locus and the mechanism through which they exert their effect have not yet been determined. In this study we focused on understanding the molecular basis underlying the observed association of SNPs located at the 5′ region of SORL1 with LOAD. In an attempt to link the current genetic and expression evidence, we examined the effect of the 5′ associated region on the regulation of SORL1 RNA expression that includes levels of total SORL1 mRNA, and the novel splice variant at the 5′ region of the gene that lacks exon 2(Grear et al., 2009). Building on the successful approach with SNCA(Linnertz et al., 2009) and SCARB1(Chiba-Falek et al.) studies, the current study was carried out using two human brain structures frontal cortex and temporal cortex that are differentially vulnerable to AD pathology. Tissue was obtained from post mortem matched brain tissues of unaffected individuals, thus we can directly assess the genetic contribution to the regulation of SORL1 expression, avoiding other confounding factors arising from the neurodegeneration associated with AD.
2. Methods
2.1. Brain samples
Brain tissue samples, including frontal cortex (n=92), and a subset of matched temporal cortex (n=52), from neurologically healthy controls (Table 1) were obtained through the Kathleen Price Bryan Brain Bank (KPBBB) at Duke University, the Brain and Tissue Bank for developmental Disorders at the University of Maryland, the Layton Aging & Alzheimer’s Disease Center at Oregon Health and Science University, and the National NeuroAIDS Tissue Consortium (NNTC). The ethnicity of all samples was white and all post mortem interval (PMI) were <24 hours. Demographics for these samples are included in Table 1.
Table 1.
Demographic description of the brain samples
| FC | TC | |
|---|---|---|
| Ntotal | 92 | 52 |
| Nmale | 55 | 29 |
| Nfemale | 37 | 23 |
| Age mean±SD | 74.1±16.5 | 77.6±14.1 |
| PMI mean±SD | 14.3 ± 9.6 | 11.2±7.7 |
| White % | 100 | 100 |
FC- frontal cortex, TC-temporal cortex,
PMI- post mortem interval.
2.2. DNA extraction and genotyping
Genomic DNA was extracted from brain tissues by the standard Qiagen protocol. Genotypes within the human genome region of interest (chr 11: 120,834,164-120,898,894) were determined by mining previously published genome-wide genotyping database that was performed using Illumina Human Hap550K chips(Heinzen et al., 2008). For samples not in the database genotypes determination of each Single Nucleotide Polymorphism (SNP) was performed by allelic discrimination using TaqMan SNP Genotyping Assays (Applied Biosystems, Foster City, CA) using the ABI 7900HT and following the manufacture’s protocol. All genotypes were tested for Hardy-Weinberg Equilibrium (Table 2).
Table 2.
The analyzed SNPs at SORL1 5′ region
| rs Number | Map position(bp) |
Distance from previous |
SNP type |
Orig. order(Rogaeva et al., 2007) |
MAF | HW (p) |
FC total (p1a,p2b) |
FC delta2 (p1a,p2b) |
TC total (p) |
TC delta2 (p) |
|---|---|---|---|---|---|---|---|---|---|---|
| rs661057 | 120834164 | Intron1 | 4 | 0.413 | 0.98 | 0.97,0.65 | 0.38,0.23 | 0.59 | 0.16 | |
| rs4936632 | 120840938 | 6774 | Intron1 | 0.473 | 0.42 | 0.81,0.67 | 0.12,0.11 | 0.07 | 0.32 | |
| rs7945931 | 120846472 | 5534 | Intron2 | 0.098 | 0.39 | 0.52,0.78 | 0.33,0.45 | 0.008 | 0.79 | |
| rs11218301 | 120847297 | 825 | Intron2 | 0.418 | 0.28 | 0.27,0.87 | 0.13,0.11 | 0.52 | 0.34 | |
| rs11600875 | 120862178 | 14881 | Intron3 | 0.092 | 0.32 | 0.16,0.87 | 0.005,0.003 | 0.44 | 0.46 | |
| rs4631890 | 120863571 | 1393 | Intron3 | 0.495 | 1.00 | 0.83,0.88 | 0.83,0.90 | 0.22 | 0.50 | |
| rs676759 | 120864475 | 904 | Intron4 | 0.364 | 0.52 | 0.42,0.28 | 0.76,0.44 | 0.35 | 0.16 | |
| rs2298525 | 120866225 | 1750 | Intron5 | 0.109 | 1.00 | 0.14,0.40 | 0.26,0.33 | 0.01 | 0.86 | |
| rs4935775 | 120894712 | 28487 | Intron8 | 11 | 0.451 | 0.38 | 0.65,0.53 | 0.1,0.14 | 0.77 | 0.27 |
| rs2298813 | 120898894 | 4182 | T528A | 13 | 0.065 | 0.64 | 0.43,0.42 | 0.32,0.33 | 0.11 | 0.57 |
FC, frontal cortex; TC, temporal cortex; total, total SORL1-mRNA; delta2, delta-2-SORL1-mRNA.
p value for the entire sample size, n=91;
p value when only matched sample were included in the analysis, n=51.
2.3. RNA extraction and cDNA synthesis
Total RNA was extracted from brain samples (100 mg) using TRIzol reagent (Invitrogen, Carlsbad, CA) followed by purification with an RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. RNA concentration was determined spectrophotometrically at 260nm, while the quality of the purification was determined by 260nm/280nm ratio that showed values between 1.9 and 2.1, indicating high RNA quality. Additionally, quality of sample and lack of significant degradation products was confirmed on an Agilent Bioanalyzer. For all samples that were included in the subsequent analysis the RNA Integrity Number (RIN) measurements were greater than 7 validating the RNA quality control. Next, cDNA was synthesized using MultiScribe RT enzyme (Applied Biosystems, Foster City, CA) under the following conditions: 10 min at 25 °C and 120 min at 37 °C.
2.4. Real time PCR
Real-time PCR was used to quantify the levels of human SORL1 mRNAs, overall (total-SORL1) and the alternative splice variant lacking exon 2 (delta-2-SORL1). Briefly, duplicates of each sample were assayed by relative quantitative real-time PCR using the ABI 7900HT for analysis of the level of total-SORL1 and delta-2-SORL1 messages as compared in brain tissues to mRNAs encoding the human neuronal proteins Enolase 2 (ENO2) and Synaptophysin (SYP). ABI MGB probe and primer set assays were used to amplify the target cDNAs, total-SORL1 (ID Hs00268342_m1, 72bp) and delta-2-SORL1 (custom assay, 111bp); and the two RNA reference controls, ENO2 (ID Hs00157360_m1, 77bp) and SYP (ID Hs00300531_m1, 63bp) (Applied Biosystems, Foster City, CA). The custom assay to detect specifically the alternative splicing variant, delta-2-SORL1 was designed to target the novel exons 1 and 3 junction. Each cDNA (10 ng) was amplified in duplicate in at least two independent runs (overall ≥ 4 repeats), using TaqMan Universal PCR master mix reagent (Applied Biosystems, Foster City, CA) and the following conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles: 15 sec at 95 °C, and 1 min at 60°C. As a negative control for the specificity of the amplification, we used RNA control samples that were not converted to cDNA (no-RT) and no-cDNA/RNA samples (no-template) in each plate. No observable amplification was detected. Data were analyzed with a threshold set in the linear range of amplification. The cycle number at which any particular sample crossed that threshold (Ct) was then used to determine fold difference, whereas the geometric mean of the two control genes served as a reference for normalization. Fold difference was calculated as 2−ΔΔCt; ΔCt=[Ct(target)-Ct (reference)]. ΔΔCt =[ΔCt(sample)]-[ΔCt(calibrator)]. The calibrator was a particular brain RNA sample used repeatedly in each plate for normalization within and across runs.
For assay validation we generated standard curves for SORL1, delta-2-SORL1 and each reference assay, ENO2 and SYP using different amounts of human brain total RNA (0.1-100 ng). In addition, the slope of the relative efficiency plot for SORL1 and for delta-2-SORL1 with each internal control (ENO2 or SYP) was determined to validate the assays. The slope in the relative efficiency plot for SORL1, delta-2-SORL1 and the reference genes were <0.1, showing a standard value required for the validation of the relative quantitative method.
2.5. Statistical analysis
All analyses were carried out using SAS statistical software, Version 9.1 (SAS Institutes, Cary, NC). SORL1 mRNA and delta-2-SORL1 mRNA fold expression values of each sample were analyzed repetitively and the results of all replicates were averaged. All average values were expressed as mean±SE. Tissues comparisons were done by paired 2 tailed T-tests. Correlations of SORL1 genotypes with expression traits (fold levels of total SORL1 mRNA and delta-2-SORL1 mRNA) were assessed using the Generalized Linear Models procedure (PROC GLM). A log transformation (log2) was performed on all mRNA levels to assure normal distribution (Bengtsson et al., 2005). SNP genotypes were coded as a dominant model pooling homozygous for minor alleles and heterozygous genotypes, as homozygosity for the minor allele was rare for most SNPs. All models controlled for gender, age, PMI, and tissue source.
Linkage disequilibrium (r2) within the studied 5′ region of SORL1 was determined using Haploview software (version 4.2) (Barrett et al., 2005).
3. Results
3.1. The effect of secondary (non-genetic) variables on SORL1-mRNA level
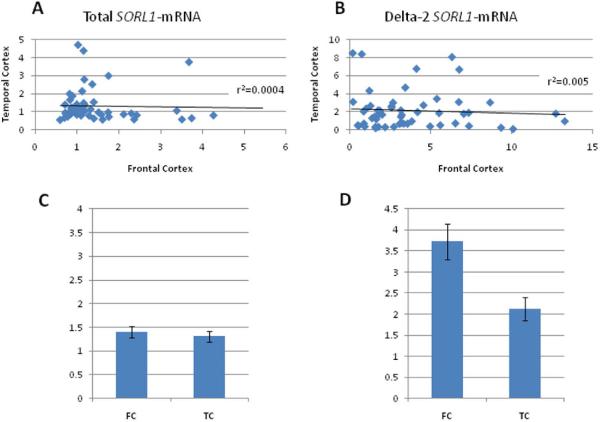
Recently, two novel SORL1 splice variants were identified one of which, in frame exon 2 skipping, was more abundant (represented 1-5% of total mRNA)(Grear et al., 2009). Using RT-PCR for amplification from exon1 through exon 3 followed by direct sequencing of the shorter product detected on a gel we confirmed the generation of SORL1-mRNA isoform lacking exon 2 in human brain (Supplementary Material 1 and Supplementary Fig1). In order to understand the regulation of SORL1 expression we focused on the major mRNA isoforms, i.e. the full length message and the relatively abundant, delta exon 2 splicing variant. SORL1-mRNAs fold levels [total SORL1/(SYP, ENO); delta-2-SORL1/(SYP, ENO)] were measured in 144 brain tissue samples obtained from 92 subjects (all white, 60% males; Table 1). One sample was removed as an outlier from the association analyses. First, we assessed the correlations with confounding factors that might affect RNA levels. No significant associations of total SORL1 and delta-2-SORL1 mRNAs levels were observed in temporal (n= 51) and frontal cortex (n= 91) with sex, age, or post mortem interval (PMI). Tissue source was significantly associated with the expression levels of frontal cortex overall and splice variant (p<0.0001 and p=0.0004, respectively). All of the subsequent analyses were adjusted for tissue source, sex, age and PMI. Next, the effect of specificity of the brain tissue region on SORL1 expression was assessed. To carry out this analysis we used matched samples, i.e. samples of the two different brain structures obtained from the same cadaver (n=51). No correlations in SORL1 mRNAs expression were found between frontal and temporal cortexes for total (r2=0.0004; Fig.1A) or delta-2 splice form, (r2=0.005; Fig. 1B). Frontal cortex showed significantly higher average levels of delta-2-SORL1 mRNA compared with temporal cortex (p=0.003). The average delta-2-SORL1 mRNA levels observed in the frontal cortex samples was nearly two fold higher compared to the average delta-2-SORL1 mRNA levels detected in the temporal cortex (Fig. 1D). On the contrary, comparison of the average total SORL1-mRNA fold levels between matched samples of temporal and frontal cortexes revealed no significant differences (p= 0.53, Fig. 1C).
Figure 1. Comparison of SORL1 RNA levels between matched brain regions obtained from the same subjects (n=51).
Fold levels of total (A, C) and delta-2 (B, D) -SORL1-mRNAs were assayed by real-time RT-PCR using TaqMan technology and calculated relative to the geometric mean of SYP- and ENO-mRNAs reference control using the 2−ΔΔCt method. The analysis included matched frontal (FC) cortex and temporal cortex (TC) samples obtained from 51 subjects. (A) Total SORL1-mRNA levels in FC were not correlated with total SORL1-mRNA levels in TC (r2=0.0004). (B)Delta-2 SORL1-mRNA levels in FC were not correlated with delta-2 SORL1-mRNA levels in TC (r2=0.005). (C and D) The bar graph presents the average of SORL1-mRNA s fold expression (mean±SE) of the 51 subjects for each brain region. (C) No significant differences in the levels of total SORL1-mRNA levels were found between FC and TC (p= 0.53). (D) delta-2-SORL1 mRNA levels were significantly higher in the FC compared with TC (p=0.003).
3.2. Defining the genomic region of interest
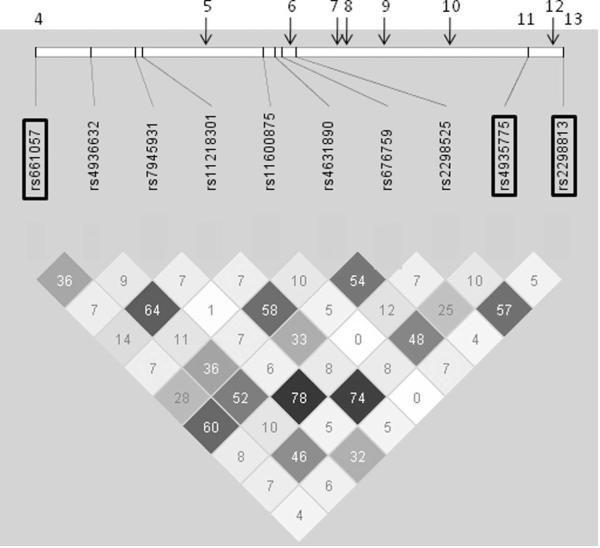
In the present work we focused on genetic functional consequences that are primarily controlled by transcription and/or splicing mechanisms. The new identified splicing event, exon 2 exclusion, was included within the SORL1 5′ LOAD- associated region. Therefore, we hypothesized that our putative expression regulatory variants are most likely positioned within the 5′ region of the gene. Based on the reproducible LOAD associations reported within the putative regulatory genomic region(Bettens et al., 2008; Cellini et al., 2009; Lee et al., 2008; Lee et al., 2007; Li et al., 2008; Meng et al., 2007; Rogaeva et al., 2007; Tan et al., 2007), we narrowed down the analysis to contain only the region between the original SNP4 (rs661057; chr 11: 120,834,164) to SNP13 (rs2298813; chr 11: 120,898,894)5, encompassing the broader SORL1 5′genomic region reported to be associated with LOAD(Bettens et al., 2008; Cellini et al., 2009; Lee et al., 2008; Lee et al., 2007; Li et al., 2008; Meng et al., 2007; Rogaeva et al., 2007; Tan et al., 2007). Ten tagging SNPs in the Illumina Human Hap550K chip were mapped within the region of interest (Table 2 and Fig. 2).
Figure 2. Linkage disequilibrium (LD) structure of the 5′ region of SORL1 gene.
Genotypes of ten SNPs from the total sample set of this study (N=92) were used to determine LD (r2) within the SORL1 5′ region using Haploview software. r2 values are shown within cells. Black cells, strong LD; shaded of grey cells, intermediate; white cells evidence of recombination. SNPs that were included in the original LOAD association study(Rogaeva et al., 2007) are marked in rectangles. Upper panel- the relative positions of the original SNPs that were not analyzed in this study are designated in arrows and the SNPs are marked in numbers according to the original order5.
3.3. The effect of genotypes on total SORL1-mRNA expression levels in human brain tissues
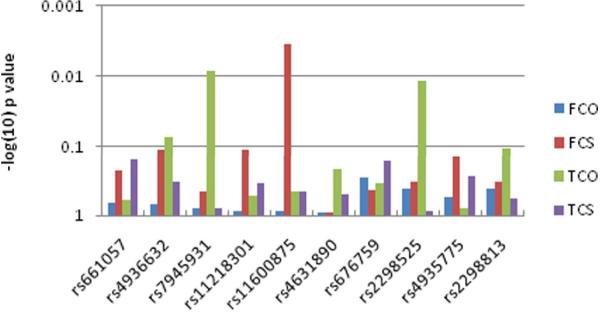
We tested the effect of variants in the 5′ region of SORL1 on total SORL1-mRNA levels with ten tagging SNPs (Table 2). Two SNPs, rs7945931 and rs2298525, showed a significant effect on total SORL1-mRNA expression in the temporal cortex (p=0.008 and 0.01; Fig. 3). These two SNPs are in high LD (r2=0.78), hence likely tagging a common genetic effector. In the temporal cortex carriers of the SNP rs7945931 minor allele, ‘A’ (AA and AG; n=10), showed higher average expression levels of total SORL1 mRNA than the non-carriers (GG, n= 41), amounting to a nearly two fold increase (Fig. 4). A similar effect in the temporal cortex was observed with SNP rs2298525, where carriers of the minor allele ‘A’ had increased overall levels of SORL1-mRNA (data not shown). In the frontal cortex, on the contrary, no association was identified between total SORL1-mRNA and the 5′ region genotypes, neither in the analyses of the entire frontal cortex samples set (n=91), nor when constrained to matched samples only (n=51) (Fig. 3 and Table 2).
Figure 3. Correlations of SORL1 genotypes with expression traits.
Fold levels of total SORL1 mRNA and delta-2-SORL1 mRNA in FC and TC were assessed using the Generalized Linear Models procedure (GLM). A log transformation (log2) was performed on all mRNA levels to assure normal distribution(Bengtsson et al., 2005). SNP genotypes (n=10) were coded as a dominant model pooling homozygous for minor alleles and heterozygous genotypes, as homozygosity for the minor allele was rare for most SNPs. The bars represent the p values.
FCO- frontal cortex, total SORL1 mRNA; FCS- frontal cortex, delta-2-SORL1 mRNA; TCO-temporal cortex, total SORL1 mRNA ; TCS- temporal cortex, delta-2-SORL1 mRNA.
Figure 4. Effect of SNP rs7945931genotypes on human total SORL1-mRNA expression levels in human temporal cortex.
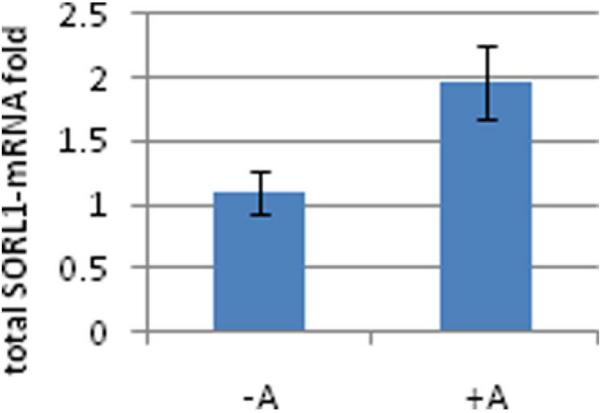
Individuals were genotyped for SNP rs7945931 and fold levels of human total SORL1-mRNA were assayed by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of SYP- and ENO- mRNAs reference control using the 2−ΔΔCt method. Analysis of the temporal cortex showed that the minor allele carriers ‘+A’ genotypes (AA, AG; n=10) correlate with significantly higher total SORL1-mRNA levels then the non carrier, ‘−A’ genotype (GG; n=41) (p=0.008). The same significant trend was observed in the analysis of SNP rs2298525 (p=0.01).
3.4. The effect of genotypes on alternative splicing of SORL1-mRNA in human brain tissues
Next, we evaluated whether SNPs at SORL1 5′ region associated with the expression levels of the novel identified splice variant, delta-2-SORL1.
Genotypes of SNP rs11600875 showed a significant effect on SORL1 exon 2 splicing levels in frontal cortex (n=91, p= 0.005, Table 2). The correlation remains significant when the analysis was limited to the matched frontal samples (n=51, p=0.003; Fig. 3 and Table 2). Carriers of the minor allele (TT and TC; n=9) had approximately two fold higher delta-2-SORL1-mRNA average levels than non carrier (CC; n= 42) (Fig. 5). The temporal cortex region, however, did not show a significant correlation with delta-2-SORL1-mRNA folds levels with any of the studied SNPs within the SORL1 5′ region (Fig. 3 and Table 2). This SNP was not in linkage disequilibrium with either of the SNPs that were associated with overall expression (Fig. 2).
Figure 5. Effect of SNP rs11600875 genotypes on human delta-2 SORL1-mRNA expression levels in human frontal cortex.
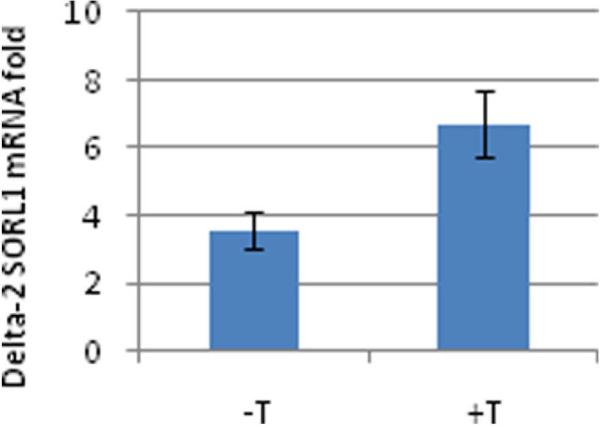
Individuals were genotyped for SNP rs11600875 and fold levels of human delta-2 SORL1-mRNA were assayed by real-time RT-PCR using TaqMan technology and calculated relative the geometric mean of SYP- and ENO- mRNAs reference control using the 2−ΔΔCt method. Analysis of the frontal cortex showed that the minor allele carriers ‘+T’ genotypes (TT, TC; n=9) correlate with significantly higher delta-2 SORL1-mRNA levels then the non carrier, ‘−T’ genotype (CC; n=42) (p=0.003).
4. Discussion
In previous studies, down regulation of SORL1 expression was reported in sporadic AD patients compared to control, particularly in neurons in the cortex and hippocampus, areas vulnerable to AD pathology (Dodson et al., 2006; Scherzer et al., 2004),(Liang et al., 2008). Analysis of afflicted brains may introduce interpretative challenges that arise from the massive cell loss, particularly neuronal loss, along with other pathologic processes accompanying neurodegeneration that may influence expression. Nevertheless, interesting associations between genetic variants and gene expression levels do not necessarily require a comparison between tissues from both cases and controls, since significant differences in gene expression levels were also shown to be associated with different genotypes in disease relevant tissues of unaffected individuals (Emilsson et al., 2008; Myers et al., 2007). In the current study we analyzed variations in SORL1 mRNA expression in the brains of unaffected people, aiming to identify such genetic expression effects in AD relevant tissue. Specifically, we looked for variations in the levels of total SORL1 mRNA and the novel exon 2 skipped transcripts and tested for association with one of the two suggested LOAD-associated regions within SORL1 locus(Bettens et al., 2008; Cellini et al., 2009; Lee et al., 2008; Lee et al., 2007; Li et al., 2008; Meng et al., 2007; Rogaeva et al., 2007; Tan et al., 2007). We hypothesized that the 5′ LOAD-associated interval of the gene might be relevant in the regulation of its RNA expression, i.e. overall levels and/or efficiency of exon 2 splicing, and therefore chose to focus on variants within the 5′ SORL1 LOAD-associated region.
Prior SORL1-LOAD association studies have described the major alleles at the 5′ SNPs as risk alleles(Bettens et al., 2008; Lee et al., 2008; Rogaeva et al., 2007; Webster et al., 2008). Although we analyzed different SNPs we looked at the same genomic interval; and in agreement to the correlation found between AD and reduced SORL1 expression, we revealed that the major alleles, presumably disease-risk alleles, correlate with lower overall SORL1 expression level. Rogaeve et al.(Rogaeva et al., 2007) also attempted to show that the LOAD associated haplotype might be attributed to some extent to lower SORL1 expression supporting the concept that reduction in SORL1 levels is likely to be a primary and pathogenic event. They were able to provide tentative support for the hypothesis that LOAD-associated haplotype in SORL1 may be associated with reduced SORL1 transcription, however the analysis was performed in lymphoblasts and was limited to the 3′ LOAD-associated block (insufficient samples to test the 5′ SNPs effect). Here for the first time we report the genetic effect of the 5′ LOAD-associated interval on the SORL1 transcription in disease relevant tissues.
In addition, we found a genetic effect on the splicing regulation of SORL1 exon 2 that was independent of the effect on overall expression. The associated SNP rs11600875 is positioned within intron 3, ~20 Kb downstream from exon 2 and a bioinformatics search predicted neither creation nor disruption of putative splicing elements (Pupasuite2, data not shown). It is unlikely that this is the actual regulatory variant, and possible serves as a marker for a proximate variant that harbors a direct effect on exon 2 splicing event. The delta-2 isoform encodes a SORL1 protein that lacks amino acids V96-D134 that contains the vacuolar protein sorting 10 (VPS10) domain. This region may be involved in amyloid precursor protein (APP) processing, but the functional consequences of the loss of this domain are not known. Biochemical experiments are needed in order to interpret the functional relevance of changes in delta-2 SORL1 levels to AD risk.
Our analysis included functionally and anatomically distinct brain regions from unaffected age-matched brains. Frontal cortex and temporal cortex are known to have differential susceptibility to AD pathology, ranging from severely afflicted temporal cortex, to the frontal cortex that is involved in a later stages of disease(Braak and Braak, 1991). Interestingly, the genetic control identified in this study was not global (across the two brain regions) but brain-region specific, indicating regionally differential regulation of SORL1-mRNAs expression in both transcriptional and splicing levels. Cuenco et al. found that distinct SORL1 variants were associated with MRI and neuropathological changes related to AD in different brain regions, supporting the hypothesis of functional importance of multiple SORL1 variants(Cuenco et al., 2008). Here we provide functional evidence to further support the importance of distinct SORL1 genetic variants in different regions of the brain. Furthermore, from a subset of subjects (n=12) hippocampus samples were available for measuring SORL1-mRNAs levels. In the hippocampus (n=12) a 50% increase in the average expression level of delta-2 SORL1-mRNA was detected with the minor allele carriers genotypes (n= 3) when compared to non carrier genotype (n=9) (Supplementary Fig. 2B). This result suggested a possible effect of the minor allele on increased levels of delta-2 SORL1-mRNA, which will need to be explored with a larger group. Interestingly this trend of the effect supports the significant genetic association observed in the frontal cortex. Using this small group of hippocampus tissues, on the other side, identified no associations between total SORL1-mRNA and the 5′ region genotypes (Supplementary Fig. 2A).
Our study examined several SNPs in two brain tissues for association with expression, and as such, is subject to false positive associations. Therefore, P values should be interpreted with caution. After Bonferroni adjustment for 20 hypotheses (10 variants × 2 tissues), the associations with all variants became non significant. However, given that these were not entirely independent hypotheses (SNPs in linkage disequilibrium), this may be an overly conservative interpretation. Moreover, the coincidental location of SORL1 SNPs associated with both expression (our study) and AD (other studies) lends support to a putative functional role of this locus that warrants further investigation in a larger cohort. Furthermore, we suggest using deep re-sequencing analysis of the expression associated region to identify potential causal variant/s that will be pursued in follow up studies, e.g. in vitro in cell based system and in vivo using animal models, and which will further our understanding of the underlying biology of associations with LOAD.
Supplementary Material
Acknowledgments
We thank the Kathleen Price Bryan Brain Bank (KPBBB) at Duke University funded by NIA AG028377, the National NeuroAIDS Tissue Consortium (NNTC), Layton Aging & Alzheimer’s Disease Center at Oregon Health and Science University funded by NIA AG008017, and the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore (NICHD contract no. HHSN275200900011C, Ref. No. NO1-HD-9-0011), for providing us with the brain tissues. We would like also to thanks Dr. Randy Woltjer, Dr. Kathleen Hayden, Dr. Lauren Warren, Mari Szymanski, and John Ervin for their assistance in obtaining the required brain samples for the study.
This work was funded in part by a pilot grant (to OC) from the Kathleen Price Bryan Alzheimer’s Disease Research Center at Duke University (funded by NIA AG028377) and the Ellison Medical Foundation New Scholar award AG-NS-0441-08 (to OC). Ms. Sunita Saith was partially funded by awards from the Trinity College Research Forum in Biology at Duke University and from the American Foundation for Aging Research.
Footnotes
Disclosure statement The authors have no actual or potential conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29:769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Cellini E, Tedde A, Bagnoli S, Pradella S, Piacentini S, Sorbi S, Nacmias B. Implication of sex and SORL1 variants in italian patients with Alzheimer disease. Arch Neurol. 2009;66:1260–1266. doi: 10.1001/archneurol.2009.101. [DOI] [PubMed] [Google Scholar]
- Chiba-Falek O, Nichols M, Suchindran S, Guyton J, Ginsburg GS, Barrett-Connor E, McCarthy JJ. Impact of gene variants on sex-specific regulation of human Scavenger receptor class B type 1 (SR-BI) expression in liver and association with lipid levels in a population-based study. BMC Med Genet. 11:9. doi: 10.1186/1471-2350-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenco KT, Green RC, Zhang J, Lunetta K, Erlich PM, Cupples LA, Farrer LA, DeCarli C. Magnetic resonance imaging traits in siblings discordant for Alzheimer disease. J Neuroimaging. 2008;18:268–275. doi: 10.1111/j.1552-6569.2007.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson SE, Gearing M, Lippa CF, Montine TJ, Levey AI, Lah JJ. LR11/SorLA expression is reduced in sporadic Alzheimer disease but not in familial Alzheimer disease. J Neuropathol Exp Neurol. 2006;65:866–872. doi: 10.1097/01.jnen.0000228205.19915.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Grear KE, Ling IF, Simpson JF, Furman JL, Simmons CR, Peterson SL, Schmitt FA, Markesbery WR, Liu Q, Crook JE, Younkin SG, Bu G, Estus S. Expression of SORL1 and a novel SORL1 splice variant in normal and Alzheimers disease brain. Mol Neurodegener. 2009;4:46. doi: 10.1186/1750-1326-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen L, Madsen P, Moestrup SK, Lund AH, Tommerup N, Nykjaer A, Sottrup-Jensen L, Gliemann J, Petersen CM. Molecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated protein. J Biol Chem. 1996;271:31379–31383. doi: 10.1074/jbc.271.49.31379. [DOI] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70:887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, Mayeux R. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O’Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, Caselli RJ, Kukull WA, McKeel D, Morris JC, Hulette CM, Schmechel D, Reiman EM, Rogers J, Stephan DA. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, Browndyke JN, Hulette CM, Welsh-Bohmer KA, Chiba-Falek O. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One. 2009;4:e7480. doi: 10.1371/journal.pone.0007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, Feng BJ, Bertoli-Avella AM, van Swieten J, Axenovich TI, Heutink P, van Broeckhoven C, Oostra BA, van Duijn CM. A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 2007;81:17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Lee JH, Cheng R, St George-Hyslop P, Mayeux R, Farrer LA. Association between SORL1 and Alzheimer’s disease in a genome-wide study. Neuroreport. 2007;18:1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Offe K, Dodson SE, Shoemaker JT, Fritz JJ, Gearing M, Levey AI, Lah JJ. The lipoprotein receptor LR11 regulates amyloid beta production and amyloid precursor protein traffic in endosomal compartments. J Neurosci. 2006;26:1596–1603. doi: 10.1523/JNEUROSCI.4946-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzer CR, Offe K, Gearing M, Rees HD, Fang G, Heilman CJ, Schaller C, Bujo H, Levey AI, Lah JJ. Loss of apolipoprotein E receptor LR11 in Alzheimer disease. Arch Neurol. 2004;61:1200–1205. doi: 10.1001/archneur.61.8.1200. [DOI] [PubMed] [Google Scholar]
- Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282:32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer’s disease in Chinese. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, Coon KD, Zismann VL, Beach T, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Huentelman MJ, Joshipura K, Walker D, Heward CB, Ravid R, Rogers J, Papassotiropoulos A, Hardy J, Reiman EM, Stephan DA. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Bujo H, Kusunoki J, Seimiya K, Kanaki T, Morisaki N, Schneider WJ, Saito Y. Elements of neural adhesion molecules and a yeast vacuolar protein sorting receptor are present in a novel mammalian low density lipoprotein receptor family member. J Biol Chem. 1996;271:24761–24768. doi: 10.1074/jbc.271.40.24761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.