Abstract
The accurate division of duplicated DNA is essential for maintenance of genomic stability in proliferating eukaryotic cells. Errors in DNA replication and chromosomal segregation may lead to cell death or genomic mutations that lead to oncogenic properties. Thus, tight regulation of DNA replication and mitosis is essential for maintaining genomic integrity. Cell division cycle 6 (Cdc6) is an essential factor for initiating DNA replication. Recent work shows that phosphorylation of Cdc6 by pololike kinase 1 (Plk1), one of the essential mitotic kinases, regulates mitotic exit mediated by Cdk1 and separase. Here we discuss how pre-replicative complex factors are connected with Plk1 and affect mitotic exit.
Key words: Plk1, Cdc6, DNA replication, mitotic exit, chromosomal segregation
Coupling Plk1 and Pre-RC Proteins
Polo-like kinase 1 (Plk1), a mammalian ortholog of Drosophila Polo, is a serine/threonine kinase that is involved in several mitotic events including centrosome maturation, bipolar spindle formation, chromatin segregation and cytokinesis.1–3 Polo kinases are highly conserved and include Plx1 in Xenopus, Polo in Drosophila and Cdc5 in budding yeast. Plk1 is highly expressed in proliferating cells. Expression of Plk1 increases in S phase and peaks during M phase.1,3 Its specific activity increases at the G2/M boundary as the result of phosphorylation.4 The multiple roles of Plk1 during the mitotic process depend on the interaction of the polo-box domain of Plk1 with its substrates for phosphorylation at different subcellular locations.5–7 Plk1 localizes to centrosomes in prophase, then is enriched in the kinetochores after nuclear envelope breakdown. Plk1 is recruited to the spindle pole in metaphase, the central spindle in anaphase and mid-body in telophase.1,5,8
To investigate the function of Plk1 at other points in the cell cycle, several groups have studied the role of Plk1 in cellular events such as DNA damage repair and DNA replication. Starting in late mitosis, a DNA pre-replicative complex (pre-RC) begins to assemble on an origin recognition complex (ORC). Initiation factors Cdc6 and Cdt1 are recruited to the ORC after which the Mcm2-7 hexamer (MCM complex) is loaded.9–11 The critical event in pre-RC formation is the activation of the MCM complex helicase at the origin of replication. Although pre-RCs form in late mitosis and G1 phase, MCM complexes remain inactive at replication origins and unwound DNA is not detected.11 Two protein kinases, the Dbf4-Cdc7 kinase (DDK) and cyclin-dependent kinase (CDK), trigger the initiation of DNA synthesis by phosphorylating the MCM complexes.
It is plausible that mitotic Plk1 and pre-RC components work together in mitosis or S phase when the Plk1 level increases. This suggestion is supported by studies that show Plk1 interacts with and phosphorylates several pre-RC proteins. Mcm2 and Mcm7 are shown to bind Plk1 through its polo-box domain in co-immunoprecipitation assays and by MALDI-mass spectrometry or yeast two-hybrid studies.12,13 Plk1 binds to Mcm7 in response to adriamycin, a topoisomerase inhibitor, that causes DNA breaks, suggesting that Plk1 may regulate the phosphorylation of Mcm7 following disruption of DNA replication.13 Immunofluorescence studies using GFP-tagged Mcm subunits showed that the MCM complex proteins weakly localize to the centrosome. The depletion of Mcm3 induces multinucleated cells and abnormal organization of microtubules.13 These data suggest that apart from the role in pre-RC formation Mcm subunits have an additional role in mitotic progression.
Further evidence for a connection between components of the pre-RC and mitosis was revealed when Orc2 was shown to be a binding partner and substrate of Plk1 in yeast two-hybrid studies and in vitro kinase assays.13 Orc2 also colocalizes to the centrosome along with Plk1.13–15 Although the physiological role of the phosphorylation of Orc2 by Plk1 remains to be elucidated, the depletion of Orc2 by siRNA affects chromosome segregation and results in multipolar spindle organization and multinucleated cells.13,14 This suggests that Plk1-mediated phosphorylation of Orc2 may function in mitotic progression. Expression of mutant Orc2 or Orc5 in Drosophila leads to arrest in metaphase with abnormally condensed chromosomes.16 As with Orc2, Orc6 also localizes to nuclear chromatin during interphase and is found at the kinetochore and centrosomes in prophase.17,18 Cells that lack Orc6 have cytokinetic defects and a multinucleated phenotype.17 Along with the localization of Orc6, the phenotype of Orc6-depleted cells indicates that Orc6 may function in chromosomal segregation and cytokinesis before DNA replication is initiated.17
Further data indicating a role for pre-RC components in mitosis are reports showing that Dbf4/Cdc7, one of the major kinases involved in pre-RC initiation, interacts with Plk1 and regulates mitotic exit by blocking Plk1 in budding yeast.19,20 The depletion of Dbf4 leads to nuclear segregation defects and misorientated spindles.20 The evidence that the deficiency of pre-RC components results in common mitotic defects suggests that pre-RC components are tightly connected with roles in mitotic progression or exit before pre-RCs form.
Cdc6-Mediated Mitotic Exit through Phosphorylation by Plk1
During pre-RC assembly ORC recruits Cdc6, which is required for loading the MCM complex, and is essential for S-phase entry.21 After replication is initiated, Cdc6 is phosphorylated by S-phase CDK, which facilitates the degradation of yeast Cdc6 or translocation of human Cdc6 from the nucleus to the cytosol. Both cases lead to downregulation of Cdc6 activity to prevent re-initiation of replication.21–23 However, there are reports that a high level of mammalian Cdc6 is still bound to chromatin in the nucleus after S phase,24–28 implicating Cdc6 in another post-S-phase function. Other evidence shows that Cdc6 may have a function in mitotic progression. Overexpression of Cdc6 causes a cell cycle delay and blocks the onset of mitosis, which is dependent on inhibition of mitotic CDK activity and the integrity of checkpoint pathways.29–31 In addition to delaying mitotic entry, Cdk1 inhibition by Cdc6 has been implicated in accelerating timely exit from mitosis through APC/Cdc20 modulation.32–35 Mouse Cdc6 has been reported to be associated with the mitotic apparatus,36 showing that Cdc6 persists in mitotic cells as does Orc6, which is associated with the outer kinetochore during mitosis.17
Recently, we have reported that Cdc6 phosphorylation by Plk1 regulates chromosomal segregation through separase and cycin-dependent kinase 1 (Cdk1) in late mitosis.37 Plk1 interacts with Cdc6 through its C-terminal polo-box domain. Cdc6 interacting with Plk1 is hyperphosphorylated and binding is elevated in mitotic cells.37 Perhaps the high levels of Cdc6 and Plk1 serve to promote their interaction. It is also possible that Cdc6 may be first phopshorylated by Cdk2 thus priming it for Plk1 phosphorylation. Immunochemistry studies reveal that Cdc6 and Plk1 colocalize to the spindle pole in metaphase and the central spindle in anaphase.37 In telophase, Cdc6 localizes in the newly formed nuclei whereas Plk1 is found at the mid-body. The colocalization of Cdc6 and Plk1 in the central spindle during anaphase may suggest an important function for p-Cdc6. Plk1 phosphorylates Cdc6 on T37 and expression of T37V mutant induces multinucleated cells and incompletely separated nuclei.37 However, cells co-expressing Cdc6-T37V and Cdc25C, which overcome mitotic delay, have a high percentage of mis-segregated chromosomes compared to cells expressing wild type Cdc6 and Cdc25C (unpublished data), suggesting that phosphorylation of Cdc6 on threonine 37 is important for mitotic exit.
A Cdc6 deficiency results in mitotic defects such as chromosomal misalignment, chromosomal lagging during segregation and multinucleated cells. The location of Cdc6 and the phenotype in Cdc6-depleted cells are very similar to those of Mcm3-, Orc2- or Orc6-depleted cells mentioned above.13,14,17,18 Orc6 shows mid-zone localization by late anaphase and begins to redistribute to the daughter nuclei by telophase,17 similar to the localization of Cdc6. Depletion of Orc6 induces defects in chromosomal segregation and multinucleation. In addition, Mcm3 and Orc2 are located in the centrosome with Plk1 and the depletion of each protein induces multinucleated cells with insufficiently organized microtubules,13 indicating that replication factors are important to regulate mitotic exit that is connected with pre-RC formation. It remains unclear how DNA replication factors can regulate chromosomal segregation and cytokinesis directly.
Inhibition of Cdk1 and Activation of Separase by pCdc6
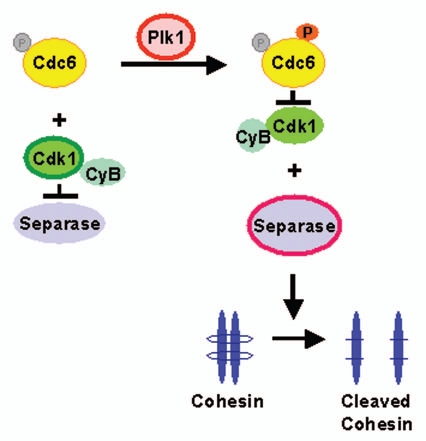
Accurate control of chromosomal separation is critical for the faithful transfer of genetic material. Defects in chromosomal segregation in somatic cells often lead to aneuploidy associated with abnormal development and tumorigenesis.38–40 Chromosomal segregation is controlled by separase activity, which cleaves cohesin, leading to sister chromosome separation.41 Premature activation of separase and chromosomal missegregation are prevented by multiple inhibitory mechanisms. One widely accepted model is that separase activity is regulated by securin through direct binding before securin is degraded by APC/C.42,43 However, other mechanisms exist to regulate separase activity because cohesin is cleaved in an appropriate cell cycle phase in securin-deficient mice.44 In addition to securin-dependent inhibition of separase, cyclin B1/Cdk1 associates with separase and inhibits its enzyme activity.45–47 Yeast Cdc6 contributes to downregulation of Cdk1 activity in mitosis and the N-terminus of Cdc6 is important for interaction with Cdk1,48 or cyclin B1.49 The phosphorylation of Cdc6 on T37 directly promotes its interaction with Cdk1 and suppresses the activity of Cdk1. Separase is then activated and Rad21, a component of cohesin, is cleaved in cells expressing wild-type Cdc6 but not mutant Cdc6, indicating that phosphorylation of Cdc6 on T37 leads to inhibition of Cdk1 through their direct interaction, which in turn leads to activation of separase.37 These results imply that phosphorylation of Cdc6 by Plk1 in mitosis regulates the activity of separase through association with Cdk1 (Fig. 1). Plk1 phosphorylates Cdc6, which binds to Cdk1 and inhibits its activity. This sequential action leads to the release and activation of separase in a securin-independent manner, to promote chromosomal segregation. The importance of inhibition of Cdk1 and cohesin cleavage in anaphase progression is described in a recent work.50 Microinjection studies reveal that cohesin cleavage and Cdk1 downregulation are sufficient to form daughter nuclei in cells arrested in metaphase,50 suggesting that cells pass from metaphase to anaphase when cohesin cleavage is combined with Cdk1 inhibition. Thus, Cdc6 could be a mediator of Cdk1 inhibition and cohesin cleavage resulting in mitotic progression to anaphase.
Figure 1.
Possible role of phosphorylated Cdc6 in separase activation. Plk1-mediated phosphorylation of Cdc6 promotes its interaction with Cdk1, resulting in the inhibition of Cdk1 and the activation of separase. Consequently, separase accelerates chromosomal segregation and the progression of anaphase.
Another securin-independent mechanism for regulating separase activity involves the phosphatase PP2ACdc55, which has been suggested to be an inhibitor of separase downstream of Shugoshin.51 Interestingly, yeast Cdc6 interacts with PP2ACdc55 in mitosis and has an inhibitory effect because cells expressing Cdc6 show downregulation of PP2ACdc55 activity in anaphase.32 These reports raise another possibility that Cdc6 binds with PP2ACdc55, disturbs the activity of PP2ACdc55, and consequently promotes separase activity in anaphase, leading to proper exit from mitosis.52 These data imply that Cdc6 regulates separase activity to promote anaphase through two different ways: by inhibiting Cdk1 and/or by inhibiting PP2ACdc55.
Thus, the functions of Cdc6 in two consecutive phases of the cell cycle converge. The first sequential role of Cdc6 along with Plk1 is to insure the fidelity of chromosome separation by promoting the regulation of separase leading to exit from mitosis. This leads to the second role of Cdc6 in assembly of pre-RC complexes in preparation for DNA replication.
Concluding Remarks
The function of many proteins is often changed by protein modification such as phosphorylation and dephosphorylation. As a result of these modifications, one protein can have several functions in a temporal- or spatial-dependent manner. Studies about the detailed modification of multiple factors will clarify the exact function of each molecule in different phases of the cell cycle.
Acknowledgments
We thank Jean Dahl and Eleanor Erikson for critical reading the manuscript. This work was supported by National Institutes of Health Grant GM 59172 and R.L.E. is the John F. Drum American Cancer Society Research Professor.
Abbreviations
- Plk1
polo-like kinase 1
- Cdc6
cell division cycle 6
- Cdk1
cyclin-dependent kinase 1
- Orc
origin recongnition complex
- Mcm
mini-chromosomal maintenance
References
- 1.Petronczki M, Lenart P, Peters JM. Polo on the Rise-from mitotic entry to cytokinesis with Plk1. Dev Cell. 2008;14:646–659. doi: 10.1016/j.devcel.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinase and oncogenesis. Oncogene. 2005;24:267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 3.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 4.Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. PNAS USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 7.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, et al. The molecular basis for phospho-dependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 8.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diffley JF. Regulation of early events in chromosome replication. Curr Biol. 2004;14:778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 10.Bell SP, Dutta A. DNA replication in eukaryotic cells. Ann Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 11.Remus D, Diffley JFX. Eukaryotic DNA replication control: Lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Tsvetkov L, Stern DF. Interaction of chromatin-associated Plk1 and Mcm7. J Biol Chem. 2005;280:11943–11947. doi: 10.1074/jbc.M413514200. [DOI] [PubMed] [Google Scholar]
- 13.Stuermer A, Hoehn K, Faul T, Auth T, Brand N, Kneissl M, et al. Mouse prereplicative complex proteins colocalise and interact with the centrosome. Eur J Cell Biol. 2007;86:37–50. doi: 10.1016/j.ejcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig JM, Earle E, Canham P, Wong LH, Anderson M, Choo KH. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum Mol Genet. 2003;12:3109–3121. doi: 10.1093/hmg/ddg330. [DOI] [PubMed] [Google Scholar]
- 16.Pflumn M, Botchan M. Development. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- 17.Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- 18.Prasanth SG, Mendez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Phil Trans R Soc Lond Biol Sci. 2004;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YC, Weinreich M. Dbf4 regulates the Cdc5 polo-like kinase through a distinct non-canonical binding interaction. J Biol Chem. 2010;285:41244–41254. doi: 10.1074/jbc.M110.155242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller CT, Gabrielse C, Chen YC, Weinreich M. Cdc7p-Dbf4p regulates mitotic exit by inhibiting polo kinase. PLoS Genetics. 2009;5:1000498. doi: 10.1371/journal.pgen.1000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- 22.Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian Cdc6 by cyclin A/Cdk2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delmolino LM, Saha P, Dutta A. Multiple mechanisms regulate subcellular localization of human CDC6. J Biol Chem. 2001;276:26947–26954. doi: 10.1074/jbc.M101870200. [DOI] [PubMed] [Google Scholar]
- 24.Coverley D, Pelizon C, Trewick S, Laskey RA. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J Cell Sci. 2000;113:1929–1938. doi: 10.1242/jcs.113.11.1929. [DOI] [PubMed] [Google Scholar]
- 25.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6 and mini-chromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 2001;20:4263–4277. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T. Nuclear organization of DNA replication initiation proteins in mammalian cells. J Biol Chem. 2002;277:10354–10361. doi: 10.1074/jbc.M111398200. [DOI] [PubMed] [Google Scholar]
- 28.Alexandrow MG, Hamlin JL. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression and overexpression of cyclin A. Mol Cell Biol. 2004;24:1614–1627. doi: 10.1128/MCB.24.4.1614-1627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.Greenwood E, Nishitani H, Nurse P. Cdc18p can block mitosis by two independent mechanisms. J Cell Sci. 1998;111:3101–3108. doi: 10.1242/jcs.111.20.3101. [DOI] [PubMed] [Google Scholar]
- 31.Clay-Farrace L, Pelizon C, Santamaria D, Pines J, Laskey RA. Human replication protein Cdc6 prevents mitosis through a checkpoint mechanism that implicates Chk1. EMBO J. 2003;22:704–712. doi: 10.1093/emboj/cdg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boronat S, Campbell JL. Mitotic Cdc6 stabilizes anaphase-promoting complex substrates by a partially Cdc28-independent mechanism, and this stabilization is suppressed by deletion of Cdc55. Mol Cell Biol. 2007;27:1158–1171. doi: 10.1128/MCB.01745-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Archambault V, Li CX, Tackett AJ, Wasch R, Chait BT, Rout MP, et al. Genetic and biochemical evaluation of the importance of Cdc6 in regulating mitotic exit. Mol Biol Cell. 2003;14:4592–4604. doi: 10.1091/mbc.E03-06-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calzada A, Sacristán M, Sánchez E, Bueno A. Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature. 2001;412:355–358. doi: 10.1038/35085610. [DOI] [PubMed] [Google Scholar]
- 35.Perkins G, Drury LS, Diffley JF. Separate SCF(CDC4) recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;20:4836–4845. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Illenye S, Heintz NH. Functional analysis of bacterial artifical; chromosomes in mammalian cells: Mouse Cdc6 is associated with the mitotic spindle apparatus. Genomics. 2004;83:66–75. doi: 10.1016/s0888-7543(03)00205-2. [DOI] [PubMed] [Google Scholar]
- 37.Yim H, Eriksn RL. Cell division cycle 6, a mitotic substrate of polo-like kinase1, regulates chromosomal segregation mediated by cyclin-dependent kinase 1 and separase. PNAS USA. 2010;107:19742–19747. doi: 10.1073/pnas.1013557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang N, Ge G, Meyer R, Sethi S, Basu D, Pradhan S, et al. Overexpression of separase induces aneuploidy and mammary tumorigenesis. PNAS USA. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumada K, Yao R, Kawaguchi T, Karasawa M, Hoshikawa Y, Ichikawa K, et al. The selective continued linkage of centromeres from mitosis to interphase in the absence of mammalian separase. J Cell Biol. 2006;172:835–846. doi: 10.1083/jcb.200511126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirth KG, Wutz G, Kudo NR, Desdouets C, Zetterberg A, Taghybeeglu S, et al. Separase: A universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol. 2006;172:847–860. doi: 10.1083/jcb.200506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- 42.Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 43.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 44.Pfleghaar K, Heubes S, Cox J, Stemmann O, Speicher MR. Securin is not required for chromosomal stability in human cells. PLoS Biol. 2005;3:416. doi: 10.1371/journal.pbio.0030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stemmann O, Gorr IH, Boos D. Anaphase topsyturvy: Cdk1 a securin, separase a CKI. Cell Cycle. 2006;5:11–13. doi: 10.4161/cc.5.1.2296. [DOI] [PubMed] [Google Scholar]
- 46.Gorr IH, Boos D, Stemmann O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol Cell. 2005;19:135–141. doi: 10.1016/j.molcel.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Ji JY, Crest J, Schubiger G. Genetic interactions between Cdk1-cyclinB and Separase complex in Drosophila. Development. 2005;132:1875–1884. doi: 10.1242/dev.01780. [DOI] [PubMed] [Google Scholar]
- 48.Elsasser S, Lou F, Wang B, Campbell JL, Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mimura S, Seki T, Tanaka S, Diffley JF. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–1123. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- 50.Oliveira RA, Hamilton RS, Pauli A, Davis I, Nasmyth K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nature Cell Biology. 2010;12:185–192. doi: 10.1038/ncb2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clift D, Bizzari F, Marston AL. Shugoshin prevents cohesin cleavage by PP2A (Cdc55)-dependent inhibition of separase. Genes and Development. 2009;23:766–780. doi: 10.1101/gad.507509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan Z, Fedorov SA, Mumby MC, Williams RS. PR48, a novel regulatory subunit of protein phosphatase 2A, interacts with Cdc6 and modulates DNA replication in human cells. Mol Cell Biol. 2000;20:1021–1029. doi: 10.1128/mcb.20.3.1021-1029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]