Figure 2.
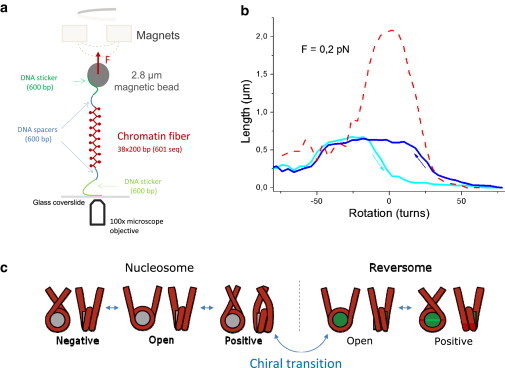
Response under topological deformation of nucleosome fibers reconstituted on 601 array. (a) Scheme of the magnetic tweezers setup. A single nucleosome/chromatosome array (∼7.6 kbp), sandwiched between two naked DNA spacers (∼600 bp each), is linked to a coated surface and to a magnetic bead. A pair of magnets placed above this molecule exerts controlled torsional and extensional constraints (17). (b) Extension-versus-rotation responses in buffer B0 (10 mM Tris, 1 mM EDTA, 0.1 mg/mL BSA) at a constant force of 0.2 pN. (Light blue) Response of a nucleosome fiber when torsion is increased from negative to positive values. In blue, response of the same fiber when torsion is decreased from positive to negative values. This cycle display a hysteresis due to the transition of individual nucleosomes to a metastable particle called a reversome. The reversome consists in the rightward wrapping of DNA around the histone octamer. This reproducible hysteresis is due to the reversible trapping of one positive turn in every reversome during chiral transition (9). (Red) Response of the corresponding naked DNA after a treatment with heparin at a concentration of 500 μg/mL. The shift in topology after nucleosome removal is consistent with the topological deformation of −1 turn per nucleosome. (c) Model of the nucleosome behavior under topological deformation. Under moderate torsion, nucleosome can adapt its topology according to the three-state model (8). Extensive positive topological deformation induces a chiral transition that leads to a reversome, in which DNA is wrapped in a rightward manner around histone octamer. This state can exist in two topological states—open or positively crossed. Topological adaptation at the nucleosomal level gives rise to a remarkable torsional elasticity of chromatin fibers compared to naked DNA (8,9).
