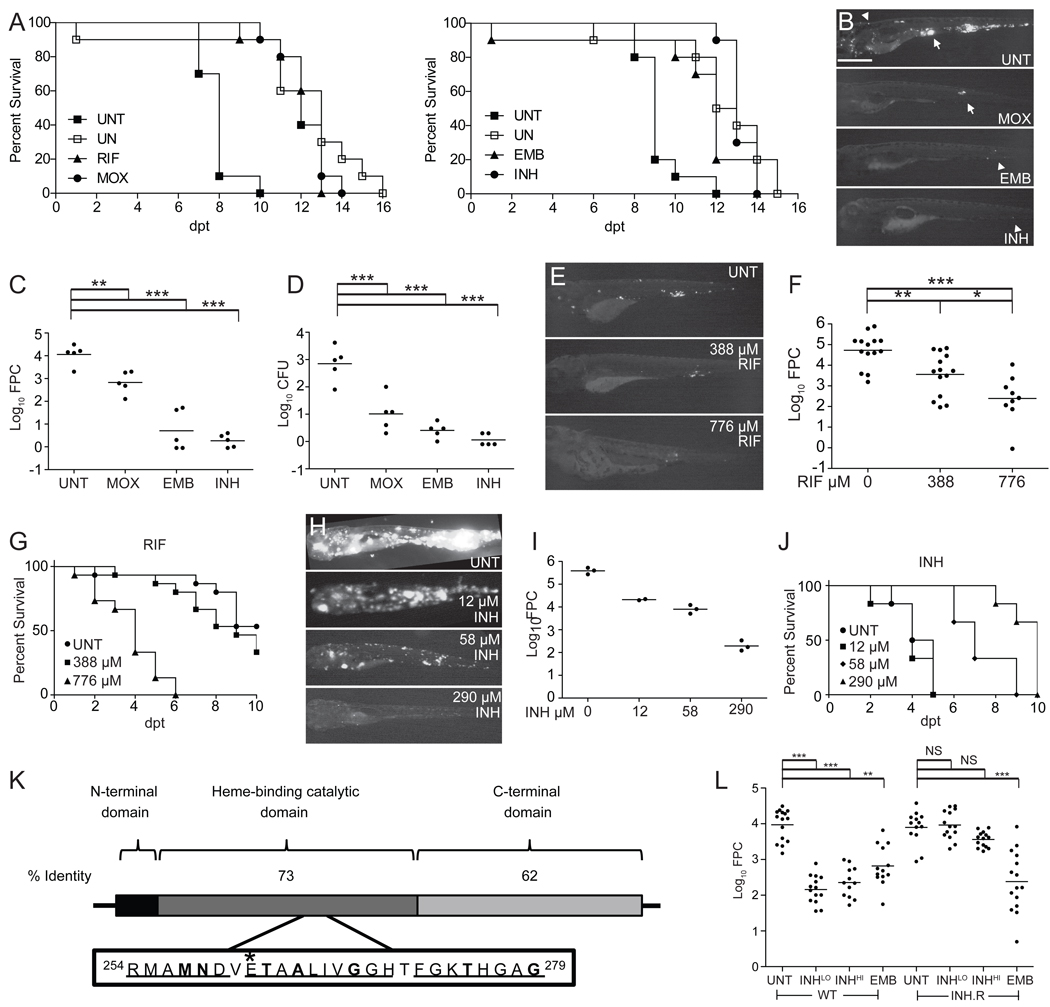
Figure 1. The Mm-larval zebrafish infection model replicates the specificity and activity of clinically relevant antitubercular drugs.
(A–D) Larvae were soaked in the MEC of RIF (388 µM), MOX (62.3 µM), EMB (1442 µM) or INH (290 µM) (Table S1). (A) Survival of uninfected (UN) larvae vs. those infected with 800 Mm and immediately treated with RIF, MOX, EMB, INH or left untreated (UNT). Left panel in the presence of 1% DMSO (see Figure S2A). Survival of treated infected larvae was significantly different from UNT larvae for all drugs (Table S2). Results are representative of at least two independent experiments.
(B–D) Larvae were infected with 155 Mm and left untreated (UNT) or treated with MOX, EMB, or INH for 4 days, prior to assessment of bacterial burdens by fluorescence microscopy (B, representative larvae are shown), FPC (C) or CFU enumeration of the lysed larvae immediately after imaging (D). Arrow, granuloma; arrowhead, single infected macrophage. Scale bar 500 µm. For C and D, individual larvae (points) and means (bars) are shown. Significance testing by one way ANOVA with Dunnett’s post test.
(E and F) Larvae infected with 46 Mm were soaked for 3 days in 388 or 776 µM RIF or left untreated. Representative fluorescence images (E) and bacterial burdens (F) of survivors are shown. Significance testing by one-way ANOVA with Tukey’s post-test.
(G) Survival of uninfected larvae upon treatment with 0, 388 or 776 µM RIF added 2 dpf. P=0.0010 for 0 vs. 776; 0 vs. 388 µM, NS by Log-rank test. N=15 per group.
(H–J) Larvae infected with 1800 Mm were left untreated or immediately soaked in 12, 58 or 290 µM INH. Representative fluorescence images (H) and bacterial burdens (I) of survivors at 4 dpt are shown. Mean ba cterial burdens (bars) compared by one-way ANOVA with Tukey’s post-test resulted in P < 0.001 for all comparisons, with the exception of 12 vs. 58 µM INH, which was not significant (P > 0.05). (J) Survival curve, N= 6 larvae per group. P=0.0018, Log-rank test for trend comparing all curves and P=0.0010 for comparison of untreated vs. 58 µM or untreated vs. 290 µM.
(K) Functional domains of KatG. Mtb KatG is 740aa and Mm KatG is 743aa. Boxed inset of Mtb catalytic domain shows regions of identity with Mm (underlined) and point mutations (in bold) that confer INH resistance in Mtb (Sandgren et al., 2009). *Position of the single amino acid substitution (E265V) in the INH-resistant Mm strain corresponds to the E261 position of Mtb KatG.
(L) Bacterial burdens of 3 dpt larvae infected with 300 WT or INH-resistant (INH.R) Mm and soaked in 290 (LO) or 2320 µM (HI) INH, 1442 µM EMB, or left untreated (UNT) beginning 1 dpi. Median log10FPC (bars) compared using Kruskal-Wallis test with Dunn's post-test. For all panels, *, P<0.05; **, P<0.01; ***, P<0.001, NS, not significant.