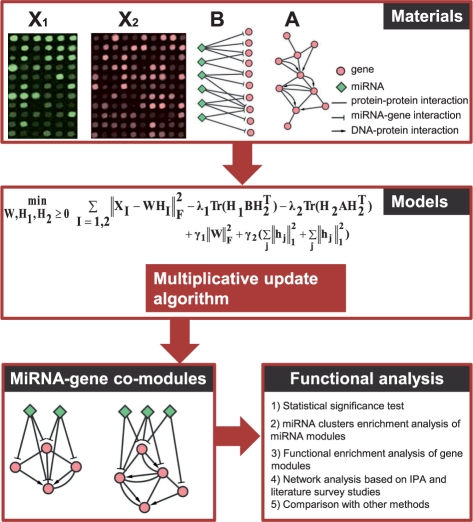
Fig. 1.
Overview of the proposed method for identifying miRNA-gene regulatory comodules. A miRNA-gene comodule is defined as the union of a set of miRNAs (a miRNA module) and a set of genes (a gene module). The inputs are (i) two sets of expression profiles (represented by the matrices X1 and X2) for miRNAs and genes, measured on the same set of samples; (ii) a gene–gene interaction network (represented by the matrix A), including protein–protein interactions and DNA–protein interactions; and (iii) a list of predicted miRNA–gene regulatory interactions (represented by the matrix B) based on sequence data. We simultaneously factor the miRNA and gene expression matrices into a common basis W and two coefficient matrices H1 and H2. At the same time, additional knowledge is incorporated into this framework with network-regularized constraints. Sparsity constraints are also imposed on this framework so as to obtain easily interpretable solutions. The decomposed matrix components provide information about miRNA-gene regulatory comodules. Then the comodules are identified based on shared components (a column in W) with significant association values in the corresponding rows of H1 and H2.