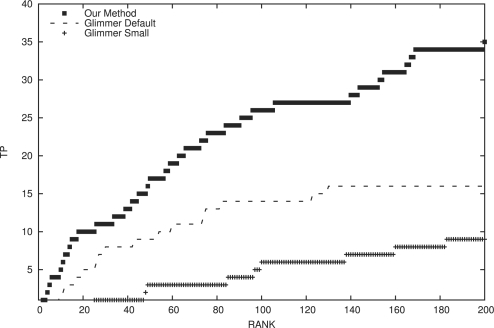
Fig. 3.
Our approach identified 35 of the experimentally validated small proteins within the top 200 predictions in our ensemble. Conversely, the default Glimmer protocol found 16 from this dataset within the top 200 predictions. Using a lower minimum size cutoff for Glimmer introduces more false positive into the ensemble, resulting in reduced performance compared with the standard Glimmer protocol. Note: results for our method in this figure are slightly poorer than the results shown in Figure 2, because this test included ORFs that overlapped the true small proteins and some overlapping ORFs scored quite well.