Figure 4. Base substitution error rates of E. coli and Plasmodium falciparum KPom1 DNA polymerase in the LacZ forward mutation assay.
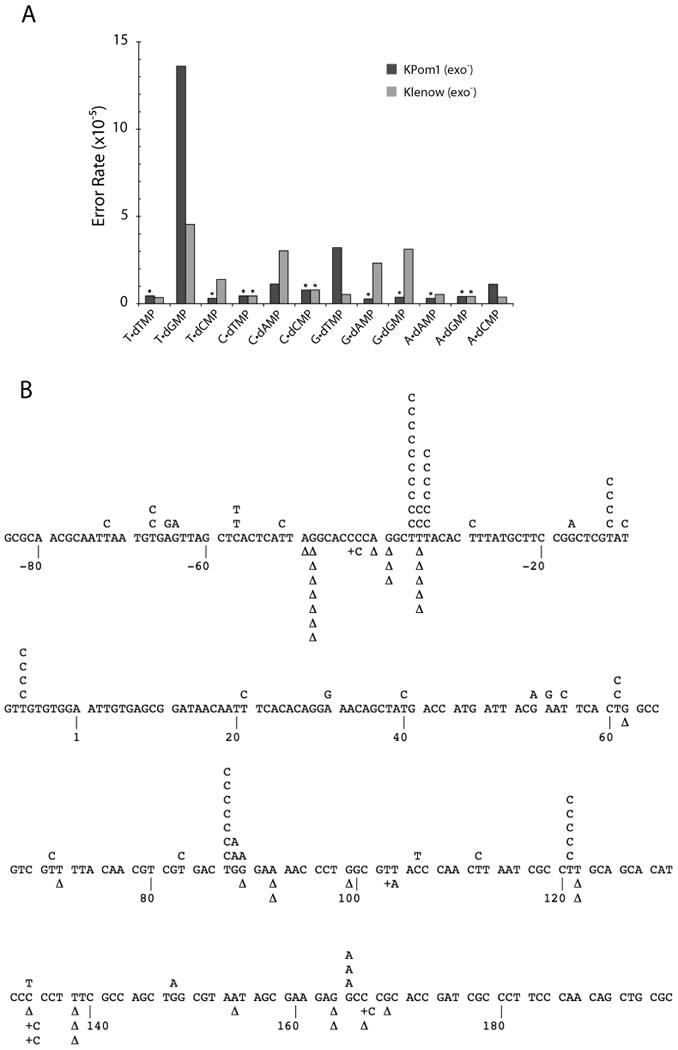
(A) Error rates for all possible base-base mispairs are shown for KPom1exo- (dark bars) and Klenowexo- (light bars). An asterisk (*) indicates less than or equal to values, due to failure to identify any mutations of a particular class. Data for Klenowexo- are from Bebenek and Kunkel 21. (B) Mutation spectrum of Kpom1exo-. The target sequence of the LacZ fragment is shown, with detected single-base substitutions indicated above the target sequence and single nucleotide deletions (Δ) and insertions (+) below.
