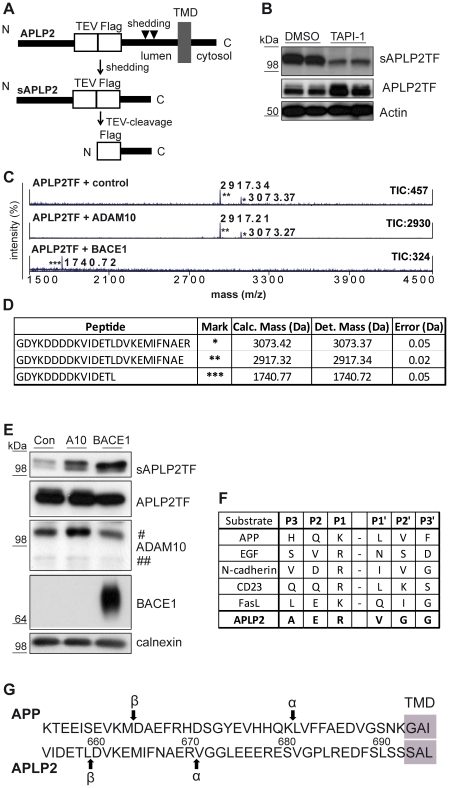
Figure 4. Mass spectrometry-based determination of APLP2 α- and β-shedding sites.
(A) Scheme of experimental procedure. A TEV-protease cleavage site followed by a FLAG-tag was introduced into APLP2 isoform 763 starting after amino acid M653 (APLP2TF). The TEV-FLAG site is positioned 39 amino acids N-terminally of the assumed start of the transmembrane domain (TMD). Shedding yields sAPLP2TF, which was immunoprecipitated and digested with TEV-protease, leading to a small peptide harboring the N-terminal FLAG-tag as well as the C-terminal cleavage site resulting from the shedding. This peptide was analyzed in a mass spectrometer. (B) HEK293 cells were transiently transfected with the APLP2TF construct and either treated with TAPI-1 (50 µM) or DMSO as a solvent control. Upon TAPI-1 treatment secreted sAPLP2TF levels were clearly decreased in the conditioned medium of the cells. An accumulation of APLP2TF levels was observed upon TAPI-1 treatment in the cell lysate. Actin was analyzed in the cell lysate as a loading control. (C) Peptides produced according to the scheme in A were obtained from HEK293 cells transiently overexpressing APLP2TF and either luciferase as a control, ADAM10 or BACE1. Mass spectrometric analysis of the peptides revealed two peaks for control and ADAM10 overexpression (*,**) while upon BACE1 overexpression only one peak with a lower m/z ratio was observed. Total Ion Counts (TIC) and centroid peak masses of the first isotopic peak are given for each isotopic peak cluster. All peaks are revealed from singly-charged peptides. (D) Determined (Det.) masses of the peaks in C (*,**,***) were compared to calculated (calc.) masses. For each mass, a corresponding peptide could be computed with less than 0.5 Da error. (E) Western Blot analysis of the samples used in C. sAPLP2TF was detected in the conditioned media, full-length APLP2 (APLP2TF), ADAM10 (A10, # immature and ## mature), BACE1 and calnexin as a loading control were detected in the cell lysates. A clear increase in sAPLP2TF levels was observed upon ADAM10 and BACE1 overexpression. sAPLP2TF has a slightly reduced apparent molecular weight upon BACE1 overexpression if compared to control (Con). (F) Summary of known ADAM10 cleavage sites in different substrates aligned with the newly detected ADAM10 cleavage site for APLP2 [41]. (G) Schematic comparison of the ectodomain shedding sites of APP and APLP2. The APP α-cleavage occurs 12 amino acids N-terminally of the transmembrane domain (TMD), for APLP2 it occurs 22 amino acids N-terminally of the TMD. For both proteins β-cleavage occurs N-terminally of an aspartate.