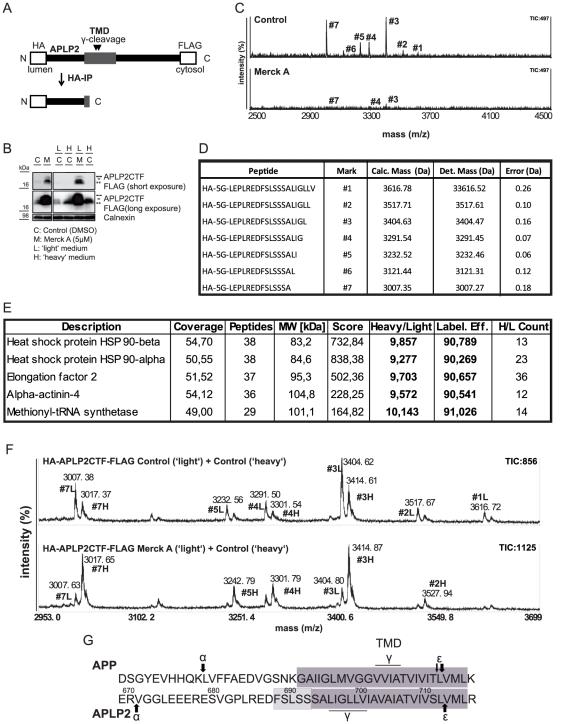
Figure 5. Mass spectrometry-based determination of APLP2 γ-cleavage site.
(A) Scheme of experimental procedure: the HA-APLP2CTF-FLAG construct was stably expressed in HEK293 cells. The construct consists of an N-terminal HA-tag (HA), a linker region consisting of five glycines (5G), the APLP2CTF sequence (APLP2) and an C-terminal FLAG-tag (FLAG). Upon γ-secretase cleavage, peptides harboring the γ-secretase cleavage site at their C-terminus were liberated. Peptides were immunoprecipitated using HA-affinity agarose and subsequently analyzed in a MALDI-TOF mass spectrometer. (B) Western Blot to confirm the γ-secretase sensitivity of the construct. Cells stably expressing HA-APLP2CTF-FLAG were treated with (M) or without (C, control) the γ-secretase inhibitor Merck A. In the presence of Merck A the CTFs accumulated in the lysate. The same was observed for cells grown in ‘light’ (L) or ‘heavy’ (H) medium. CTFs were detected with anti-FLAG antibody. Calnexin levels were analyzed as a loading control. (C+D) Mass spectrometry-based identification of the γ-secretase cleavage sites. Under control conditions, seven peaks were detected with a maximal total ion count of 497. Upon Merck A treatment, the intensity (ion count) for the peaks was reduced significantly. Peaks are labeled with identifiers #1–#7 linking them to the respective peptides sequences in the table (D). All peptides were detected with a mass error of less than 0.3 Da. (E) Determination of the labeling efficiency for the SILAC experiment. To determine the labeling efficiency, proteins from the cell lysates of the ‘heavy’ labeled cells from (F) were separated on an SDS-PAGE gel. One band was cut out and tryptic in gel digestion was performed. Peptides resulting from this digestion were analyzed in an LTQ Orbitrap Velos mass spectrometer. Proteins were identified by database search, and the labeling efficiency (Label. Eff.) was determined from the ‘heavy’ to ‘light’ ratios (Heavy/Light) of the quantifiable peptides. Further, coverage of the proteins in % (Coverage), number of unique peptides identified (Peptides), molecular weight of respective protein (MW), SEQUEST score (Score) and number of unique peptides used for quantification (H/L Count) are given. (F) Analysis of γ-secretase-dependent CTF cleavage by quantitative mass spectrometry. Upper panel: Cells without Merck A (Control) grown either in ‘light’ or in ‘heavy’ medium. Supernatants were combined, peptides immunoprecipitated and analyzed in the MALDI-TOF mass spectrometer. Peaks resulting from the ‘light’ and the ‘heavy’ labeled peptides are clearly separated (+10 Da as expected for one heavy labeled arginine). Intensities of corresponding ‘heavy’ and ‘light’ peak clusters are not identical, as the ‘heavy’ clusters are either shifted (+10 Da) or supershifted (+16 Da) due to additional incorporation of a ‘heavy’ proline resulting from arginine- to-proline conversion. Lower panel: upon treatment of the ‘light’ labeled cells with Merck A and the ‘heavy’ labeled cells with DMSO as a solvent control, the ratio between the intensity of the ‘light’ labeled peptides and the ‘heavy’ labeled peptides is significantly reduced if compared to the control experiment. This demonstrates that the identified peptide peaks are generated by γ-secretase. (G) Comparison of the position of the ε- and γ-secretase cleavage sites of APP and APLP2 in an alignment of the juxtamembrane and the transmembrane region of these two proteins. The dark boxes mark the predicted transmembrane domains (TMD), while the light box indicates amino acids which could potentially still be part of the transmembrane domain, as they do not harbor charges.