Fig. 4.
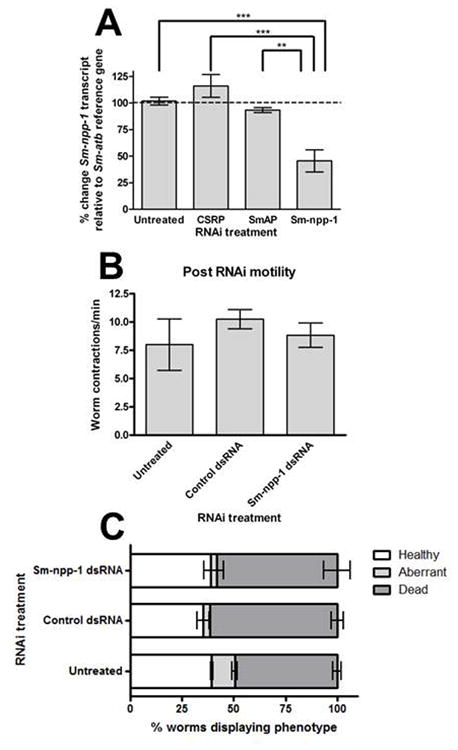
RNA interference (RNAi) analysis of Schistosoma mansoni neuropeptide precursor Sm-npp-1. (A) Quantitative PCR analysis of Sm-npp-1 transcript knockdown. Percent change in Sm-npp-1 transcript levels relative to alpha-tubulin (Sm-atb) reference gene, following treatment with one of the following double-stranded RNAs (dsRNAs): chloroplast-specific ribosomal protein (CSRP) non-target control; S. mansoni alkaline phosphatase (SmAP) non-target control; or target-specific Sm-npp-1. Knockdown of Sm-npp-1 is specific and statistically significant relative to untreated and control-treated worms. (B) Analysis of post-RNAi motility phenotype, quantified as mean number of worm whole-body contractions per minute. (C) Morphological analysis of post-RNAi phenotype. Worms were defined as one of three phenotypes, healthy (normal movement and morphology), aberrant (visible movement, aberrant morphology) or dead (no visible movement, aberrant morphology). Data points are expressed as mean % transcript change ± SEM. **P < 0.01, ***P < 0.005.
