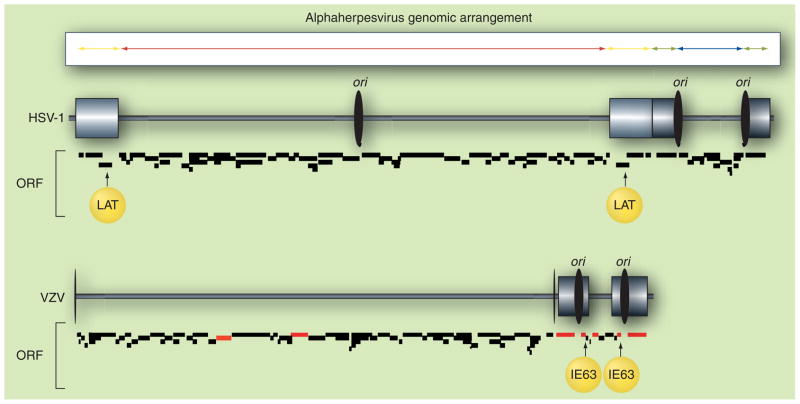
Figure 1. Herpes simplex virus type 1 and varicella zoster virus genomic structures.
The alphaherpesvirus genome is composed of two unique stretches of DNA: unique long (red) and unique short (blue). Each unique segment is covalently joined through smaller regions of inverted repeats: repeat of unique long (yellow) and repeats of unique short (green). The HSV-1 genome preserves the basic structure even though the unique regions with their attendant repeats are inverted, resulting in four isomeric forms of the virus DNA. The VZV genome lacks approximately 9 kb of DNA from each unique long repeat, which may explain why this region rarely (5%) inverts with respect to the unique short region [181]. More importantly, the unique long repeat region in HSV-1 contains the LAT locus, the major virus transcript present during latency. VZV IE63, immediate–early protein encoded by ORF 63, may fulfill some of LAT’s functions, since like LAT, ORF 63 is the most abundant and prevalent virus transcript detected during latency, and since both LAT and IE63 block virus-induced apoptosis. The red ORF in the VZV genome indicates genes whose transcripts have been cloned and sequenced from latently infected human ganglia. The ORF annotations of HSV-1 (accession number NC_001806) and VZV (accession number NC_001348) genome were supplied by PubMed. HSV-1: Herpes simplex virus type 1; LAT: Latency-associated transcript; ori: Origin of DNA replication; ORF: Open reading frame; VZV: Varicella zoster virus.