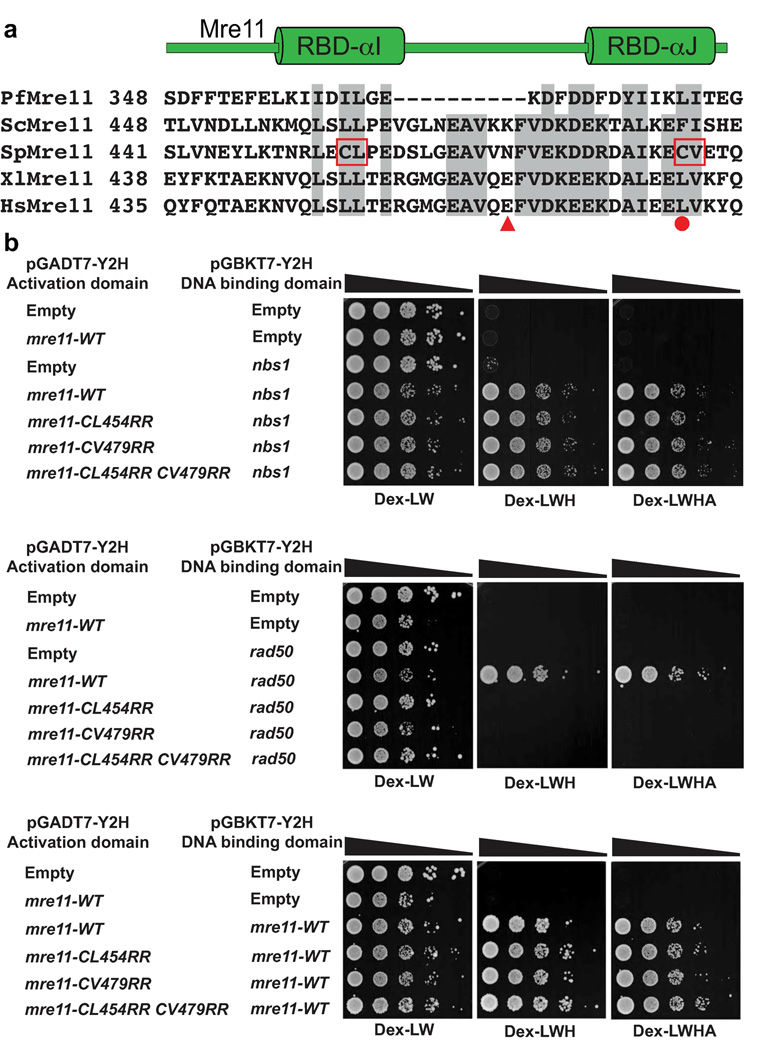
Figure 2. A conserved interface links eukaryotic Mre11 and Rad50.
(a) Multiple sequence alignment of Mre11 RBD from pfMre11, scMre11, spMre11, xlMre11 and hsMre11. Shaded regions show well-conserved residues. CL→RR and CV→RR mark hydrophobic to charged surface substitutions introduced into S. pombe Mre11 RBD. HsMre11 mutations identified in somatic colorectal cancers are highlighted (solid red circle, point mutation; triangle, truncation7). (b) S. pombe Mre11 RBD variant interactions with Rad50, Nbs1 and the Mre11 homodimeric interaction analyzed by two-hybrid. Growth on Dex-WL plates (minimal glucose media lacking tryptophan and leucine) indicates the reporter strain transformed with plasmids pGADT7 (Gal4 activating domain) and pGBKT7 (Gal4 DNA binding domain) fused to the respective proteins. Growth on Dex-LWH (less stringent; lacking histidine) and Dex-LWHA (more stringent; lacking histidine and adenine) indicates a positive two-hybrid interaction. We have previously shown that pGBKT7-mre11-WT alone does not autoactivate9 and this was not repeated here. Mre11 RBD mutants fail to interact with Rad50 yet retain homodimerization and Nbs1 interactions. Strains used are detailed in Supplementary Table 1.