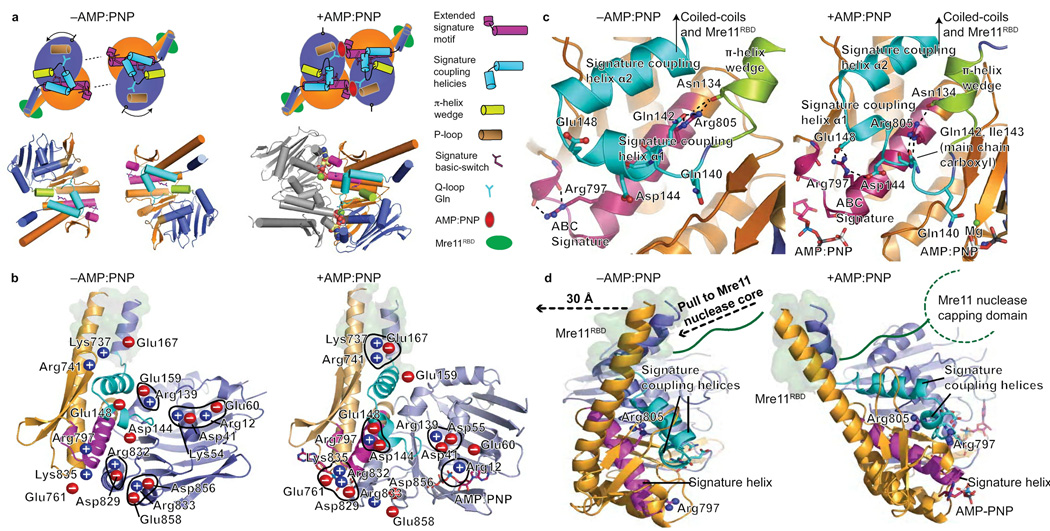
Figure 5. Rad50 nucleotide-binding induced conformational changes.
(a) Nucleotide free (−AMP:PNP) and bound (AMP:PNP) Rad50 conformations. Nucleotide binding is coordinated by the signature motif, Q-loop, P-loop, and induces a ~35° subdomain rotation. Rad50 N-lobe rotation drives the π-helix wedge into the signature coupling helices, dramatically altering signature coupling helix conformation relative to ATPase subdomain interactions. Motifs are colored as in key. (b) Twenty salt bridge switches rearrange upon nucleotide binding and coordinate domain rotations, see Supplementary Movie 1. Blue (positive) and red (negative) circles highlight charged residues. The Mre11 RBD is highlighted by a green surface representation of Rad50 residues involved in the interface. (c) Signature helix Arg797 and Arg805 rearrangements link nucleotide binding with domain rotations, conformational change of the signature-coupling helices and Q-loop, and motions in the Rad50 coiled-coils (see Supplementary Movie 2). The Mre11 RBD is highlighted as in (b). (d) Nucleotide-binding induced Rad50 ATPase C-lobe rotation relative to the N-lobe drives coiled-coil repositioning to impact bound Mre11 RBD, highlighted as in (b). The Rad50 domain rotation is transduced through coiled-coil repositioning (see Supplementary Movie 3), into a linear pull on the linker between the Mre11 RBD and nuclease capping domain as depicted by dashed arrows.