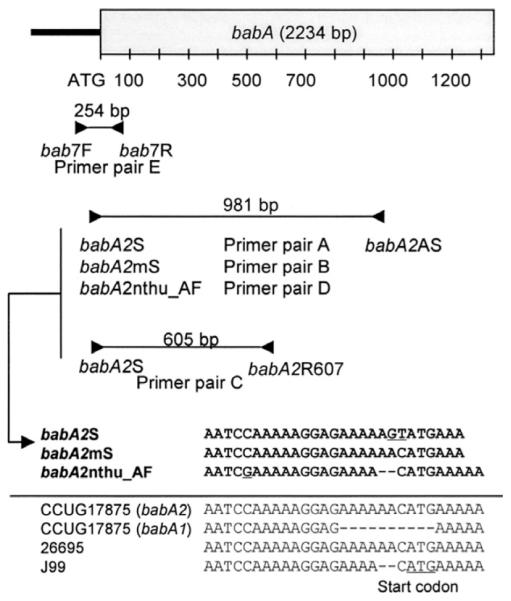
Figure 1.
Location of primer pairs used in this study. Most previous studies used primer pair A (babA2S and babA2AS); 2 nucleotides in the forward primer sequence were changed (AC to GT) from the GenBank sequence of the babA2 gene from the strain CCUG17875. The forward primer babA2mS has the same sequence arrangement as the babA2 gene from the strain CCUG17875. This primer was used in combination with the original reverse primer (primer pair B). babA2R607 is a newly designed reverse primer used in combination with the original forward primer (primer pair C). nthu_babA2F is a modified forward primer design based on GenBank sequence data from Taiwanese strains. It has 2 nucleotides deleted near the 3′ end of the original forward primer sequences. This primer was used in combination with the original reverse primer (primer pair D). The forward primer bab7F is located in the promoter region of the babA gene and was used in combination with a unique reverse primer from the 5′ region of the babA gene (bab7R) (primer pair E).