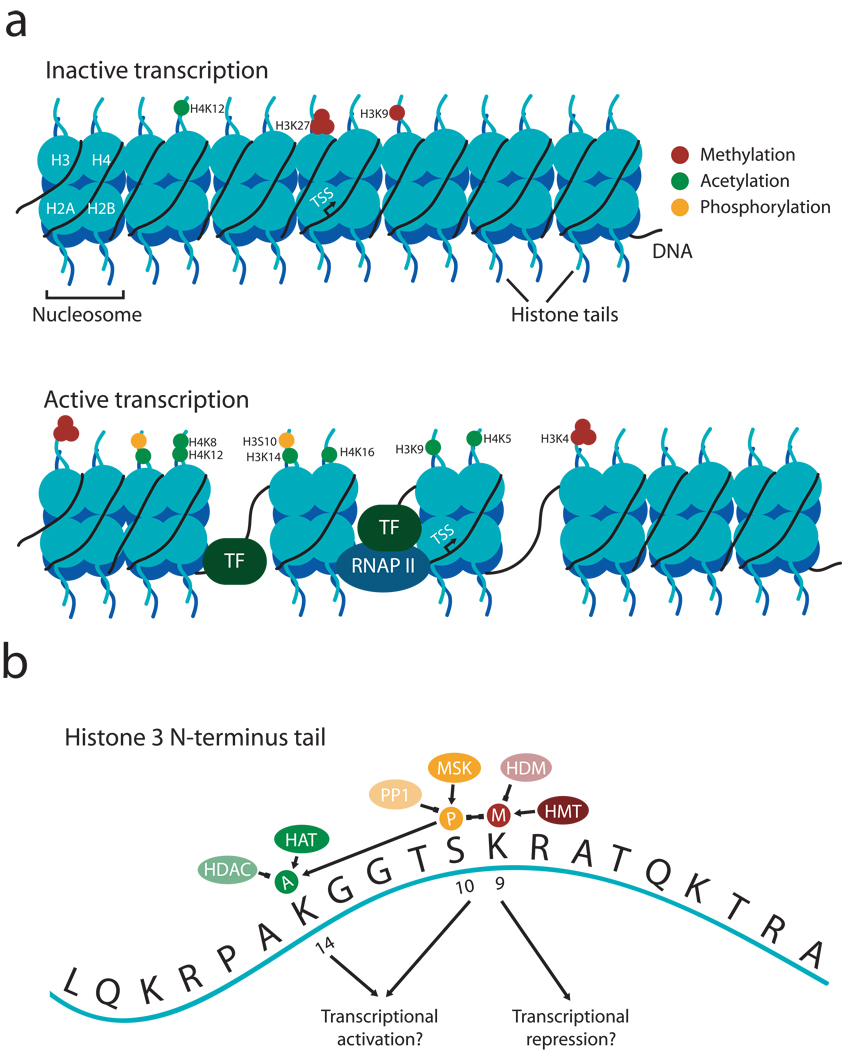
Figure 1. Dynamic regulation of histone modifications directs transcriptional activity.
A, Individual residues on histone tails undergo of a number of unique modifications, including acetylation, phosphorylation, and mono-, di-, and tri-methylation surround the transcription start site (TSS) for a given gene. These modifications in turn correlate with transcriptional repression (top), in which DNA is tightly condensed on the nucleosome and therefore inaccessible, or transcriptional activation (bottom), in which transcription factors (TF) or RNA polymerase II (RNAP II) can access the underlying DNA to promote gene expression. The specific epigenetic marks listed correlate with transcriptional activation or repression, although this list is by no means exhaustive. B, Expanded view of individual modifications on the tail of histone H3. See text for details and acronyms. The concept of a histone “code” suggests that individual marks interact with each other to form a combinatorial outcome. In this case, methylation at lysine 9 on H3 (a mark of transcriptional repression) and phosphorylation at serine 10 on H3 repress each other, whereas phosphorylation at serine 10 enhances acetylation on lysine 14 (a mark of transcriptional activation.