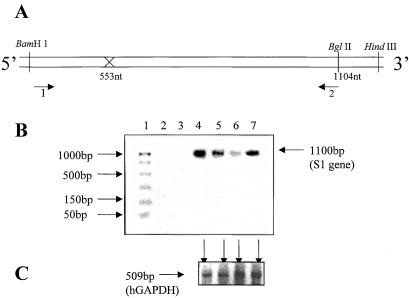
Figure 4.
To address the question of intracellular reduction of the reovirus s1 RNA by S1–553-Rz, primers were designed to amplify the full-length s1 RNA and the control hGAPDH RNA fragment (509 bases) from the same cell lysates by RT-PCR. Two terminal primers (1 and 2, A) were used to amplify the full-length s1 (1.4 kb) RNA. They had the following sequence: forward, 5′-GATCCTCGCCTACGTG and reverse, 5′-CAACGATAGATCTCCACC. Total cellular RNA was isolated from Cos-1 cells that were first transfected with 1 or 4 μg of plasmid encoding Rz-553 and then challenged with reovirus at 1 plaque-forming unit/cell. Serial dilutions of the target gene and number of cycles for PCR were optimized initially to determine the linear range. (B) Lane 1 depicts the size of standard DNAs as shown on the left. As expected the plain cells showed no evidence of the presence of s1 RNA (lane 2). The same was true if the enzyme RT was omitted from the reaction (lane 3). Reovirus infected cells, as expected, showed a prominent signal for s1 RNA (lane 4). The intensity of the signal diminished when either 1 μg (lane 5) or 4 μg (lane 6) of Rz-553 was used. Mutant-S1–553-Rz at a concentration of 4 μg failed to decrease the levels of s1 RNA (lane 7). This decrease was specific for the reovirus s1 RNA as the levels of the control RNA (hGAPDH) in all four corresponding lysates remained unchanged (C, shown by arrows).