Abstract
Neurofibromatosis type 1 (NF1) is a multi-system disease caused by mutations in the NF1 gene encoding a Ras-GAP protein, neurofibromin, which negatively regulates Ras signaling. Besides neuroectodermal malformations and tumors, the skeletal system is often affected (e.g. scoliosis and long bone dysplasia) demonstrating the importance of neurofibromin for development and maintenance of the musculoskeletal system. Here, we focus on the role of neurofibromin in skeletal muscle development. Nf1 gene inactivation in the early limb bud mesenchyme using Prx1-cre (Nf1Prx1) resulted in muscle dystrophy characterized by fibrosis, reduced number of muscle fibers and reduced muscle force. This was caused by an early defect in myogenesis affecting the terminal differentiation of myoblasts between E12.5 and E14.5. In parallel, the muscle connective tissue cells exhibited increased proliferation at E14.5 and an increase in the amount of connective tissue as early as E16.5. These changes were accompanied by excessive mitogen-activated protein kinase pathway activation. Satellite cells isolated from Nf1Prx1 mice showed normal self-renewal, but their differentiation was impaired as indicated by diminished myotube formation. Our results demonstrate a requirement of neurofibromin for muscle formation and maintenance. This previously unrecognized function of neurofibromin may contribute to the musculoskeletal problems in NF1 patients.
INTRODUCTION
Neurofibromatosis type 1 (NF1) is an autosomal genetic disease affecting multiple organ systems. Among NF1 diagnostic criteria are manifestations of neuroecodermal origin (café au lait spots, axillary freckling) and tumors (cutanous or plexiform neurofibromas, Lisch nodules, optic gliomas). Other organ systems such as the skeleton may also be affected. NF1 is caused by the loss of function mutations in the NF1 gene. It is increasingly recognized that NF1 tumors as well as other focal manifestations (e.g. tibial dysplasia) can associate with additional somatic mutations, affecting second NF1 allele (1,2). NF1 encodes neurofibromin, a large protein with a centrally positioned Ras-specific GTPase-activating domain which acts as a negative regulator of Ras/mitogen-activated protein kinase (MAPK) signaling. The function of neurofibromin was shown to be critical for the regulation of proliferation and differentiation of the hematopoietic, neural and mesenchymal lineage progenitor cells (3–5). While the molecular function of neurofibromin is best studied in the context of its tumor suppressor role, it has become clear that neurofibromin plays an essential role in embryonic development, including development of the skeletal system (6,7).
Approximately 50% of patients with NF1 suffer from skeletal manifestations (8). The most disabling skeletal manifestations are scoliosis and bowing of the long bones (tibia), the later often culminating with fractures and pseudarthrosis. In addition to the skeletal defects, reduction in muscle strength has been reported in patients with NF1 (9). A reduction in the muscle cross-section area was shown in pediatric patients with NF1 by peripheral quantitative computed tomography (pQCT) imaging, and was observed both in patients affected with bone lesions and those with no osseous manifestations (10). These data suggested that a skeletal muscle deficiency might be a feature of NF1. Since proper maintenance and function of the musculoskeletal system depends on both the skeleton and the musculature, muscular weakness may contribute to the bone phenotype in NF1.
The skeletal muscles of the limb arise from the somitic myotome. Cells at the ventrolateral edge of somites at limb levels delaminate, migrate into the limb and form the initial pre-muscle masses. These migrating and strongly proliferating pre-myoblasts express a set of characteristic marker genes, including Pax3 and Lbx1 (11,12). The migration process itself is regulated by a cascade triggered by scatter factor/hepatocyte growth factor signaling via c-met (13). The migrating pre-myoblasts themselves are committed to the myogenic lineage, but are not believed to contain positional information as to where and when to form muscles in the limb. Rather, this information seems to be conveyed by the limb mesenchyme via local paracrine signaling mechanisms (14–16). These signaling mechanisms govern the intricate interplay between proliferation and differentiation of premyogenic cells to myoblasts expressing markers for definitive myogenesis, the myogenic regulatory factors such as MyoD1 or myogenin (Myog). Myoblasts differentiate to myotubes that start to express contractile proteins such as myosin heavy chain, these myotubes subsequently fuse to form terminally differentiated multinucleated myofibers (17).
NF1 inactivation results in the embryonic lethality due to cardiac defects. Thus, conditional inactivation of the gene was required to study function in late embryogenesis and postnatal development. In order to determine how Nf1 inactivation impacts skeletal muscle development, we took advantage of Nf1Prx1 mice in which the Nf1 gene is inactivated in the limb bud mesenchyme. In these mice, Nf1 was previously shown to be inactivated in the mesenchymal cells of the developing limbs, resulting in an inactivation in osteoblasts, chondrocytes and endothelial cells (6). In addition, we found that the migrating muscle progenitor cells also undergo efficient Nf1 gene inactivation by Prx1-Cre rendering the Nf1Prx1 mice an ideal model to analyze embryonic muscle development. Here we show that Nf1Prx1 mice display a primary defect in muscle development at the level of the differentiation of myoblasts to myotubes, ultimately leading to muscular dystrophy and fibrosis.
RESULTS
Inactivation of Nf1 in the limb musculature of Nf1Prx1 mice
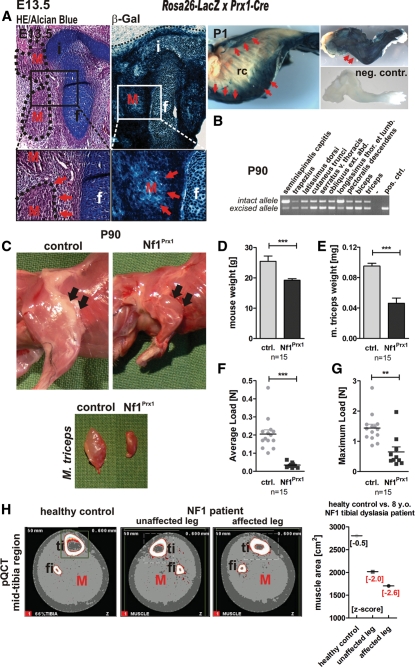
The Prx1-Cre transgene was previously shown to efficiently recombine Nf1 in mesenchymal cells in the developing limbs of Nf1Prx1 mice (6). To assess recombination in the myogenic lineage, Cre-recombinase expression and efficacy were investigated at various developmental stages using the Rosa26-LacZ reporter line. The LacZ staining revealed Cre recombinase expression throughout the entire limb at E13.5 with a defined border of expression at the proximal edge of the limb bud (Fig. 1A, left panel, dotted line). The staining was equally intensive in the cartilage and muscle primordia, indicating that migrating muscle progenitor cells express Cre recombinase while entering the limb bud in Nf1Prx1 mice. Cre expression appeared even broader in the postnatal stages (Fig. 1A, right panel), where apart from limb musculature also parts of the chest, neck and abdominal musculature were LacZ positive. A varying degree of recombination was observed in neck muscles (m. seminispinalis capitis, m. trapezius), spinal column attached muscles (m. latissimus dorsi, m. longissimus thoraci), as well as abdominal muscles (m. cutaneus trunci, m. serratus v. thoracis and m. obliquus externus abdominis) in the adult mice (Fig. 1B). Thus, at postnatal stages, Nf1Prx1 mice show Nf1 inactivation also in muscle groups affecting body posture.
Figure 1.
NF1 inactivation results in reduction of muscle size and diminished muscular force. (A) HE/Alcian blue-stained transverse sections of the hip joint region at E13.5 (left panel) and the corresponding sections of Rosa26-LacZ reporter-visualized Cre-activity in Prx1-Cre mice. Whole-mount preparations of P1 Rosa26-LacZ reporter × Prx1-Cre pups (right panel), thorax (middle panel), forelimb (upper image) and negative control—forelimb of Cre-negative littermate (lower image); rc, rib cage. Blue staining indicates where the Cre-mediated recombination event removed the stop cassette within the transgene and led to LacZ reporter expression. The border of the LacZ staining at the edge of the limb bud is demarcated (dotted line). Cre recombinase expression in the hindlimb at E13.5 is visible in muscles (M, red arrows), the primordium of the iliac bone (i) and femur (f) and in connective tissue. Robust staining is also visible in all muscle groups of the forelimb (right upper panel, red arrows point out the triceps muscle). Apart from the limb musculature also muscles of the chest, coetaneous muscles of the abdomen and parts of spinal musculature express Cre recombinase (right lower panel, red arrows). (B) PCR-based detection of the recombined allele in the adult Nf1Prx1 animal. (C) Appearance of the upper limb musculature in the adult Nf1Prx1 animal (3 months old—P90), and reduced size of the triceps muscle (arrows). (D) Reduced body weight and (E) weight of the triceps muscle in the 3-month-old Nf1Prx1 mice. Nf1-deficient mice show reduction in the grip strength measured with the force gauge device. The grip force is given in Newtons and separate graphs indicate average force load (F) and maximal force load results (G). (H) Reduced muscle cross-sectional area at the 66% tibial site (light grey shading—red M) in an 8-year-old individual with NF1 affected with unilateral tibial dysplasia. Right panel: muscle cross-sectional area measured with pQCT in both the affected and unaffected extremity of the NF1 individual, and in the extremity of a healthy age-matched control (red arrows). Note, muscle area in both the tibial dysplasia-affected leg (z-score: −2.6) and unaffected leg (z-score: −2.0) is reduced compared with the pQCT images of a healthy age-matched control and as referred to the baseline of 20 age-matched controls.
Reduced muscle mass in Nf1Prx1 mice and in the NF1 tibial dysplasia patients
As previously described, Nf1Prx1 mice exhibit short-limbed dwarfism (6). A considerable size reduction in the entire limb was observed, including reduced muscle size (Fig. 1C). While the weight of the adult Nf1Prx1 mice was on average reduced by 25% (Fig. 1D), the weight of the triceps muscle was reduced by >50% in mutant mice compared with control littermates (Fig. 1E), indicating a specific defect of muscle development. Consistently, Nf1Prx1 mice showed a dramatic reduction in the muscle force measured in a force gauge pull test. The average load of mutant mice was 0.034 ± 0.004 N versus 0.205 ± 0.023 N in controls, and the maximum load for mutant animals was 0.649 ± 0.166 N versus 1.439 ± 0.125 N in controls (Fig. 1F and G). The muscle mass reduction in Nf1Prx1 mice correlates with observations in patients as a reduction in muscle cross-sectional area has been observed in the mid-tibial region of children with NF1 (10). An example of muscle area reduction in a patient with NF1 is presented in Figure 1H, documenting the case of an 8-year-old male patient affected with tibial dysplasia. Both tibial dysplasia-affected and non-affected limbs showed reduced muscle area (z-score: −2.6/−2.0, respectively), despite the child being fully mobile and active (data not shown).
Muscle dystrophy in Nf1Prx1 mice
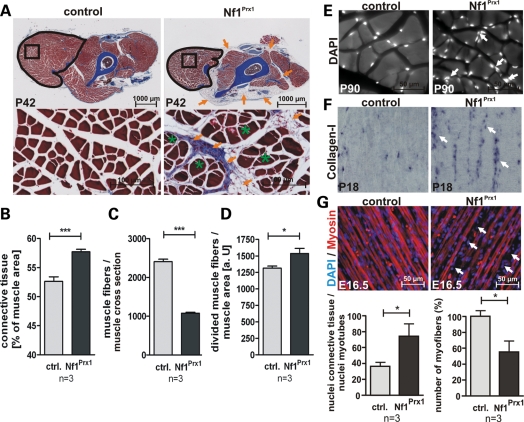
Histological analysis of the musculature in the extremities of adult Nf1Prx1 mice revealed a generalized muscle fibrosis, characterized by expansion of the collagen-rich (blue, azan stain) connective tissue (Fig. 2A and B), and a reduced total number of muscle fibers (50% reduced in the triceps) (Fig. 2C). Additionally, large areas of the dystrophic mutant musculature were also occupied by fat tissue (Fig. 2A and B, orange arrows). Mutant muscles showed a 20% increase in the number of fibers bearing cleft-like invaginations (split fibers), suggesting a disturbance of myotube architecture. Additionally, muscle fiber size appeared more variable in mutant muscles then in controls (Fig. 2D). However, no centrally positioned nuclei were observed in muscle fibers, indicating absence of overt muscle regeneration (Fig. 2E). These findings were corroborated by in situ hybridization (ISH) analysis of the P18 muscles, showing a higher abundance of collagen-I-positive connective tissue cells in the mutant muscles when compared with controls (Fig. 2F). An increase in connective tissue was already detectable in muscles of late gestation (E16.5) Nf1Prx1 embryos, which showed a higher number of cell nuclei positioned between the muscle fibers than in controls (Fig. 2G). In addition, muscle fibers were already thinned out at this stage (Fig. 2G), suggesting an early developmental origin of the muscle dystrophy and fibrosis.
Figure 2.
Loss of Nf1 results in muscle dystrophy and muscle fibrosis. (A) Histology of transverse sections of forelimbs of P42 control and Nf1Prx1 mice stained with Azan. The reduction in muscle size is visible on the overview micrographs (upper panel). Abundant connective tissue and fat tissue are marked with orange arrows. Histomorphometric measurements (B) reveal an increased area of connective tissue per muscle area, (C) reduction in muscle fiber number per muscle area and (D) an increase in the occurrence of muscle fibers with divided appearance (containing a cleft-like invaginations—green stars) in the mutant muscles when compared with controls. (E) DAPI-stained transverse sections of the adult mouse triceps muscle. Muscle fibrosis in the mutant is seen as accumulation of nuclei in the fascia region (arrows). The myofibril nuclei appear positioned normally, giving no indication of a deranged regeneration process. (F) Connective tissue labeled by in situ hybridization with a collagen-I-specific riboprobe on sagittal sections of P18 forelimb musculature. Note increased Col1a1 staining in the mutant (white arrows), reflecting expansion of the connective tissue between muscle fibers. (G) Sagittal sections of E16.5 hindlimb muscles with immunlabeled myosin and DAPI-stained nuclei. Note an abundance of nuclei outside the myosin-positive area (arrows) indicating muscle fibrosis. Quantification (below) shows accumulation of interfiber (connective tissue) nuclei relative to nuclei within myotubes and beginning rarefication of muscle fibers.
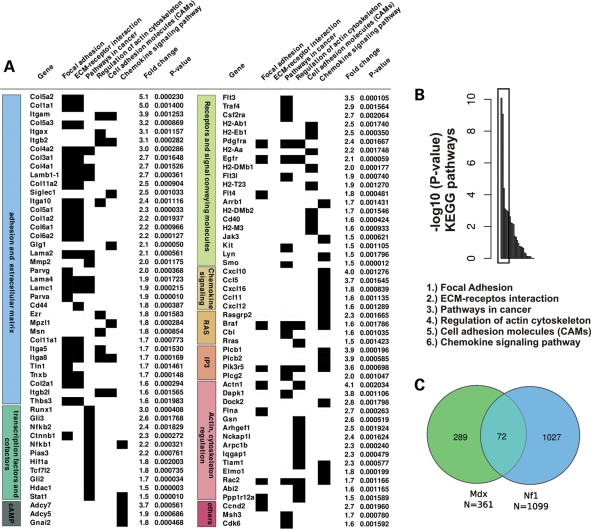
To gain insight into the molecular changes of the observed muscle dystrophy and fibrosis and to compare these with other known muscle dystrophies, we performed transcriptional profiling of the entire triceps muscles of 3-month-old wild-type (WT) and mutant animals using Affymetrix high-density microrrays (Fig. 3). Gene expression profiling was done for four mutant and four WT animals, and the significant gene regulations determined with one-way analysis of variance (ANOVA) analysis. In total, 1508 genes were found significantly (ANOVA F-factor ≥ 20) deregulated in mutant muscles when compared with controls (874 upregulated and 634 downregulated). The upregulated genes were subjected to KEGG classification and gene class enrichment analysis (see Materials and Methods). Six functional KEGG classes were found to be most significantly enriched in the data set: (i) focal adhesion, (ii) Extracellular matrix interaction, (iii) pathways in cancer, (iv) regulation of actin cytoskeleton, (v) cell adhesion molecules, (vi) chemokine signaling pathways (Fig. 3B).
Figure 3.
Dystrophic gene expression fingerprint in Nf1Prx1 mice. (A) Transcriptional fingerprint of the Nf1 muscle fibrosis. Upregulated are genes encoding extracellular matrix proteins, proteins regulating actin cytoskeleton, transcription factors involved in cancer process, receptors and signal conveying molecules involved in the regulation of MAPK, IP3 and cAMP-signaling pathways. P-values were determined with one-way ANOVA analysis. (B) Analysis of the microarray expression profile of the adult Nf1Prx1 mice triceps muscles (see Materials and Methods for KEGG class enrichment analysis). Among 1379 genes which were found upregulated in mutant muscle, six functional KEGG classes are significantly overrepresented in the data set (lower panel). (C) Overlap between gene sets deregulated in Nf1Prx1 muscles, and a gene set deregulated in muscle of mice carrying a mutation in the dystrophin gene (Mdx) (18).
Consistent with the observed muscle fibrosis, upregulated genes included extracellular matrix genes: collagens (I, II, III, IV, V, VI and XI), several glycoproteins, including thrombospondin and tenascin, as well as various integrins.
Among the upregulated transcripts were various collagens and lamins, cell adhesion molecules, like integrins, as well as various cell surface receptors, including platelet-derived growth factor receptors and epidermal growth factor receptor (Fig. 3A). Upregulated transcription factors included Runx1, Gli2 and Gli3, Catnnb1, Nfkb1 and Nfkb2. Genes involved in four signaling pathways: Ras, cAMP, PI3 and chemokine signaling pathways, were represented among enriched transcripts. A proportion of transcripts deregulated in NF1-deficient muscle encoded proteins involved in regulation of actin cytoskeleton dynamics. This group included actin itself, the actin-binding proteins, like filamin A and gelsolin, as well as modulators of rho family, small GTP binding protein signaling TIAM1 and ELMO1 (Fig. 3A).
To determine whether the dystrophic muscle phenotype correlated with features of other known muscle dystrophies, we compared transcriptional profiles of muscles of Nf1Prx1 and Mdx mice, in which muscle dystrophy is caused by a mutation in the dystrophin gene (18). The analysis revealed a considerable overlap of the two data sets (72 common gene regulations out of 361 Mdx-specific and 1099 NF1-Prx1-specific gene regulations; see Supplementary Material, Table S2); Fisher's exact test: P-value = 1.1e − 11 (Fig. 3C). Similarities included upregulation of genes encoding extracellular matrix components: collagens III, V and VI, fibulin-7, biglican and matrix metallopeptidase 2, likely representing a universal feature of the muscle fibrosis. Genes involved in cytoskeleton organization: microtubule-associated protein Tau, tubulin beta-5, ezrin, coronin and vimentin were also commonly deregulated in both data sets. Additionally, in line with what was previously reported to be a common feature of several muscle dystrophies (19), Nf1-deficient muscles showed downregulation of multiple genes encoding key mitochondrial energy production and glucose metabolism regulating proteins (Supplementary Material, Table S3). Among them were mitochondrial transcription factors A and B1, mitochondrial ribosomal proteins L2, L9, L44, L53 and S5, mito-ribosomal GTPase (Mtg1), complex IV assembly protein (COX11), complex I subunit 1 alpha (NDUFAF4), glucokinase, glutaminase 2 and tafazzin, mutations of which cause a spectrum of cardiac and skeletal muscle symptoms in Barth syndrome (20). Interestingly, we also detected marked upregulation of the gene encoding uncoupling protein 2, encoding a protein that can uncouple ATP production from mitochondrial respiration (21). Thus, like other muscle dystrophies, NF1 skeletal muscle dystrophy appears to be accompanied by a general bioenergetics crisis (19). Collectively, these data indicate that Nf1Prx1 muscle dystrophy represents a distinct pathological condition, which shares some general transcriptional properties with other known muscular dystrophies.
Muscle progenitor cell migration, proliferation and differentiation in Nf1Prx1 mice
Our initial observations prompted us to analyze whether a primary defect in embryonic myogenesis in Nf1Prx1 mice might contribute to the dystrophic phenotype observed later. Whole-mount in situ hybridization (WISH) for Lbx1 revealed no overt changes of early muscle progenitor cell migration in the Nf1-deficient limb buds at E10.5 and E11.5 (Supplementary Material, Fig. S1). Similarly, no changes were observed in the expression domain and intensity of Pax3, which together with Lbx1 labels migrating pre-myoblasts (Supplementary Material, Fig. S1).
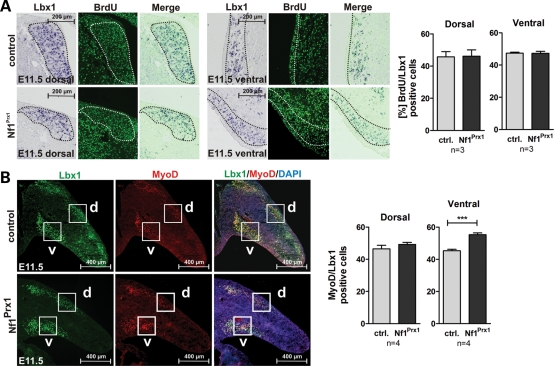
The initial pace of muscle formation in the limb is determined by the regulation of progenitor proliferation versus their differentiation to MyoD-positive myoblasts. First, we determined the proliferation rate of migrating Lbx1-positive cells at E11.5 by co-labeling for Pax3 mRNA and bromdesoxyuridin (BrdU). No changes were observed in dorsal or ventral muscle masses (Fig. 4A). To analyze the differentiation of progenitors to myoblasts, we performed immunohistochemical co-staining for Lbx1 and MyoD as specific markers for both populations. The MyoD/Lbx1 ratio was unaltered in the dorsal muscle masses in limb buds of mutant mice (Fig. 4B). However, we detected an increased MyoD/Lbx1 ratio in the ventral muscle massses (Fig. 4B), where MyoD-positive cells were also more abundant (data no shown). Altogether, this result suggested a normal migration and proliferation of pre-muscle cells in Nf1Prx1 mutants; it also suggested an increased differentiation of myoblasts in ventral muscle masses.
Figure 4.
Muscle progenitor proliferation and myoblast differentiation in Nf1Prx1 mice at E11.5. (A) Co-labeling for Lbx1 (in situ hybridization) and BrdU (antibody staining) on sections of E11.5 embryos. Proliferation index was assessed by bromodeoxyuridine (BrdU) labeling for 1h prior to sacrifice. Quantification of cells in the encircled areas is shown right for dorsal and ventral muscle masses, respectively. No differences in proliferation of Lbx1-positive cells were detected in Nf1Prx1 mutants. (B) Immunodetection of Lbx1 (green) and MyoD (red) expressing cells on the transverse sections of the forelimb buds identified migrating muscle progenitor cells and myoblasts, respectively. (B) Quantification of myoblast differentiation assessed by the relation of MyoD- versus Lbx1-positive cells. The ratio of MyoD-positive to Lbx1-positive cells is increased in the ventral region in mutant mice, which is not caused by the decrease in Lbx1-positive cells but rather by the more abundant MyoD-positive cells (data not shown). The increase appears to reflect a premature induction of the myogenic differentiation of the migrating progenitor cells in the ventral limb bud region.
Defect of muscle cell differentiation in Nf1Prx1 mice
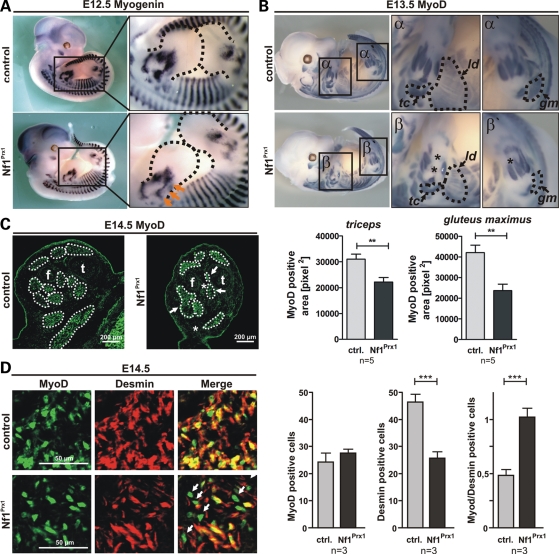
Next we aimed at determining the time point at which the first disturbances in muscle formation became apparent. MyoD and myogenin are two master regulators of skeletal muscle development and remain expressed during muscle differentiation. MyoD becomes activated in muscle progenitor cells inducing their differentiation to myoblasts, whereas myogenin is required for their terminal differentiation. Analysis of myogenin expression by WISHs indicated a slight reduction in myogenin expression in E12.5 Nf1Prx1 limb buds, suggesting an early inhibition of myoblast terminal differentiation (Fig. 5A). One day later, at E13.5, a defect in muscle formation became apparent. We used WISH for MyoD, which provides a good resolution of the forming muscle fibers, to illustrate muscle development. Specific muscle primordia were significantly reduced in size or were entirely missing (Fig. 5B, upper panel). Quantification of the MyoD-positive area showed a ∼30% reduction in the m. triceps (tc) size and a ∼50% reduction in the m. gluteus maximus (gm) size in Nf1Prx1 mice when compared with controls (Fig. 5B, lower panel, see also magnifications α′ and β′). Similarly, the m. latissimus dorsi (ld) appeared smaller and showed a rarefaction of muscle fibers in Nf1Prx1 mice (Fig. 5; magnifications α and β). The distal muscle groups in the extremities appeared most strongly affected, and some muscles were missing completely (stars in magnifications β and β′). Absence of distal muscle groups in the extremities was confirmed on transverse sections of E14.5 hindlimbs labeled with anti-MyoD antibodies (Fig. 5C) and was also observed on histological sections of postnatal stages (data not shown), indicating a continuous myogenic defect rather than a developmental delay. Together, these data indicate that the muscle differentiation process is disturbed in Nf1Prx1 mice. To assess the rate of myoblast differentiation to myotubes, we performed co-immunolabeling for MyoD, which is expressed in pre-myoblasts, myoblasts and myotubes, and the intermediate filament protein desmin, which is expressed in differentiating myoblasts and myotubes. The amount of MyoD-positive cells was equal in WT as well as in Nf1Prx1 mice (Fig. 5D). Conversely, in Nf1Prx1 mice, there was a drastic decrease in desmin-positive cells per area. Moreover, cells expressing MyoD, but not desmin were more abundant (Fig. 5D, arrows), also reflected by a MyoD/Desmin-positive cell ratio shifted towards the MyoD population. This indicates a severe disruption of myoblast terminal differentiation that might explain the rarefaction of muscles observed in Nf1Prx1 mice.
Figure 5.
Defect of myoblast differentiation in the Nf1Prx1 mice. (A and B) WISH for Myogenin and MyoD at E12.5 and E13.5. Dotted outline shows area of upper and lower limb bud. Five embryos of each genotype were analysed per stage. (A) Representative overview micrographs (left panel), magnification of the limb bud area (right panel, dashed line). Reduced expression of myogenin in the limb buds of mutant mice at E12.5 (arrows). (B) MyoD was used to label muscle areas in whole-mount embryos at E13.5. Surface of muscle primordia was determined with help of AxioVision4 -AutMess software (Zeiss). The size of most developing muscles appeared reduced. Below: quantification of the MyoD-positive area demarcating the primordial triceps (tc) and gluteus maximus (gm) muscles. Note also a profound decrease in the area occupied by the latissimus dorsi muscle (ld; marked with dotted line and arrow). Some distal muscle groups appear to be missing (stars). (C) Immunodetection of MyoD on the transverse sections thought E14.5 hindlimbs demarcating muscle areas (f, fibula; t, tibia). Note reduction in individual muscle sizes (arrows) as well as the absence of muscles (stars). (D) Determination of myoblast terminal differentiation in the ventral muscle groups by co-immunolabeling for MyoD and desmin. Note an increased amount of MyoD single positive early myoblasts in Nf1Prx1 muscles (arrows). Right panel: quantification of MyoD- or desmin-positive nuclei per area and relation of MyoD/desmin-positive nuclei show a shift towards the MyoD population and hence diminished terminally differentiated desmin-positive myoblasts.
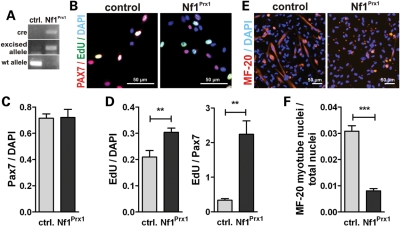
As indicated above, muscle cell differentiation in the limb is controlled by cell-autonomous and non-cell-autonomous mechanisms. To get further insight into the pathology, we used muscle satellite cells from WT and Nf1Prx1 mice as a cell culture system for myoblast differentiation. Real time quantitative PCR (RT–PCR) confirmed efficient recombination of the Nf1 locus in satellite cells from Nf1Prx1 mice (Fig. 6A). Pax7+ cell numbers were unaltered in Nf1Prx1 cultures after 3 days compared with WT, indicating normal self-renewal of satellite cells in Nf1Prx1 mice (Fig. 6B and D). Proliferation measured by incorporation of 5-ethynyl-2′-deoxyuridine (EdU) was overall increased in Nf1Prx1 cultures. This was even more pronounced when EdU labeling was assessed in Pax7+ cells (Fig. 6E). After switching to differentiation medium, WT satellite cells showed robust myogenesis as assessed by MF-20 labeling for sarcomeric myosin. Differentiation was severely decreased in Nf1-deficient satellite cells (Fig. 6C and F), confirming an intrinsic defect in muscle progenitor differentiation upon Nf1 ablation.
Figure 6.
Reduced in vitro differentiation of NF1Prx1 satellite cells. (A) RT–PCR shows efficient recombination of the Nf1flox allele in Nf1Prx1 satellite cells. (B) Three-day cultures were stained for Pax7 (marks muscle satellite cells) and EdU for proliferating cells. (C) Quantification of Pax7 cells relative to total cell number (DAPI). (D) Proliferating cells relative to total cell number (DAPI) and relative to Pax7+ cell number. (E) Muscle differentiation: 3 days after myogenic induction, myotube differentiation was analyzed with antibodies against meromyosin MF-20. (F) Quantification of the nuclei within MF20-positive area versus total nuclei (fusion index) shows reduced myogenesis in Nf1Prx1 satellite cells.
MAPK activation in Nf1-deficient muscles
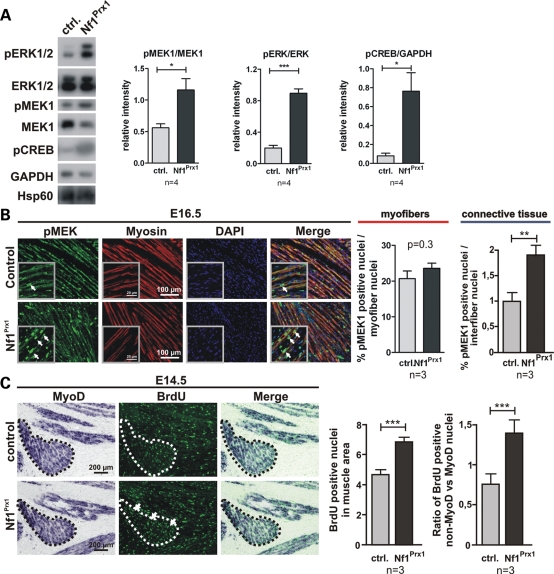
MAPK signaling, which is downstream of Ras, is known to be involved in the regulation of myogenic differentiation (22). In addition, Ras/MAPK signaling is one of the positive regulators of the cell cycle. Since we found Nf1 inactivation to result in defective myogenesis leading to dystrophic and fibrotic muscles, we analysed the MAPK pathway activation status in muscles of the Nf1Prx1 mice. First, we assessed the phosphorylation status of signaling components known to be downstream of Ras/Nf1, namely Erk1/2, MEK1 and cAMP responsive element binding (CREB) in muscles of adult mice. We detected a significant increase in pERK1/2, pMEK1 and pCREB levels in the lysates of adult Nf1Prx1 triceps when compared with WT controls (Fig. 7A). To determine whether MAPK signaling was also affected at earlier developmental stages, we performed immunohistochemical detection of pMEK1 on paraffin sections of E16.5 limbs.
Figure 7.
Hyper-activation of Ras signaling in the Nf1-deficient muscles. (A) Western blot analysis of MAPK signaling in adult mice (P90). MEK1, ERK1/2 and CREB are hyperphosphorylated in the mutant muscles. Blots probed with antibodies against pMEK1, pERK1/2 and pCREB were quantified densitometricaly (right). (B) Immunohistochemical detection of pMEK1 on longitudinal sections of the triceps muscle at E16.5. Magnified area is shown in the inserts. A robust pMEK1 signal was detected in the Nf1Prx1 muscles in the myosin-stained myofibers as well as in muscle connective tissue cells (arrows). Quantification of the MEK1-positive nuclei in the myofibers and interfiber space (right panel). WT values were set as 1. (C) Combined in situ hybridization for MyoD and immunolabeling for BrdU (1 h pulse) showing increased proliferation (BrdU, green) of cells in the MyoD-positive area (dotted line) at E14.5 (right panel). This was carried mainly by non-MyoD-positive connective tissue cells, which showed a higher proliferation index than MyoD-positive myoblasts (right).
While the level of pMEK1 within myofiber nuclei was not significantly increased, we detected a significant increase in pMEK1 levels in the interfiber nuclei representing muscle connective tissue fibroblasts in Nf1Prx1 mice (Fig. 7B).
The muscle connective tissue is strongly involved in the control of muscle differentiation and patterning, but under dystrophic conditions expands and thus contributes to fibrosis. To analyze whether this might apply to the Nf1Prx1 mouse model, we analyzed BrdU labeling in muscles at E14.5. First, we counted overall BrdU-positive cells in the muscle area as determined by ISH for MyoD. This showed that the overall proliferation index is higher in Nf1Prx1 mice (Fig. 7C). We then counted BrdU-positive nuclei in defined areas that were positive or negative for MyoD expression (MyoDpos: myoblasts or myotubes; MyoDneg: muscle connective tissue) and calculated the ratio of both cell populations. This revealed an increase in proliferating muscle connective tissue cells versus muscle cells.
DISCUSSION
The data presented here suggest that neurofibromin is required for proper formation and maintenance of skeletal muscles. Inactivation of Nf1 in the developing heart muscle has previously been shown to cause cardiac muscle fibrosis, hypertrophy and a progressive cardiac muscle dysfunction (23). Nf1 knock-out in the embryonic heart was associated with increased levels of pERK1/2, pGSK3b, pAKT and pmTOR. We now show that Nf1 is not only required for normal cardiac muscle development but is also needed during skeletal muscle formation. The terminal differentiation of myoblasts was severely impaired in the absence of Nf1 as inferred from normal muscle progenitor cell migration and proliferation at E11.5 (Fig. 4, Supplementary Material, Fig. S1), and normal differentiation of Lbx1-positive progenitors to MyoD-positive myoblasts with only a minor acceleration of differentiation in the ventral pre-muscle mass (Fig. 4). Impaired terminal myoblast differentiation was first indicated at E12.5 showing lower expression of the terminal differentiation inducer myogenin. A clear reduction in muscle size became apparent at E13.5 and E14.5, caused by a defect in myoblast terminal differentiation (Fig. 5). In vitro differentiation of satellite cells from Nf1Prx1 mice was also severely impaired, while proliferation was increased. This indicates that at least a part of the muscle phenotype observed in Nf1Prx1 mice is caused by a cell-autonomous defect. Furthermore, this indicates that in the absence of Nf1, myoblasts might fail to exit the cell cycle, which is a prerequisite for terminal differentiation. However, such an increase in myoblast proliferation was not detected in the Nf1Prx1 mice.
The increased proliferation that we detected in the muscle connective tissue cells within the muscle primordium as early as E14.5 (Fig. 6C) likely signifies the beginning of muscle fibrosis that becomes first visible in the form of expanding muscle connective tissue at E16.5 (Fig. 2G). This indicates that expansion of connective tissue is a primary defect caused by Nf1 inactivation contributing to muscle dystrophy, unlike in other known dystrophies (e.g. Duchenne muscular dystrophy caused by inactivation of the dystrophin gene), where fibrosis is a consequence of muscle degeneration. Thus, Nf1Prx1 mice develop a specific form of muscular dystrophy, which shares basic molecular features with Mdx mice, the model for Duchenne muscular dystrophy (18,24).
The observed expansion of the connective tissue within Nf1-deficient muscles was reflected by increased expression of extracellular matrix and adhesion molecules, including multiple collagens (I, II, III, IV, V, VI and XI), glycoproteins, including thrombospondin and tenascin, as well as various integrins. We also detected increased expression of Runx1, a transcription factor known to protect disused myofibers against myofibrillar disorganization and autophagy (25). Among the transcription factors upregulated in the Nf1Prx1 muscles were Gli2 and Gli3, the downstream targets of Sonic hedgehog involved in muscle development through regulation of Myf5 expression. Moreover, expression of two NFKb factors, one of which was previously implicated in the genesis of the muscle disuse induced atrophy (26) was also enhanced. Apart from deregulation of MAPK pathway genes (e.g. Ras-GRP2, Braf and Cbl), we also detected upregulation of some genes intimately involved in the IP3 pathway signaling: Plc, Plcg and Pik3r5. Interestingly, two adenylate cyclase genes Adcy7 and Adcy5 as well as the Gnai2 gene, encoding guanine nucleotide-binding protein subunit 2, involved in the hormonal regulation of adenylate cyclases, were also overexpressed indicating that cAMP signaling might be deregulated in NF1-deficient muscles. In line with this, we detected elevated levels of the cAMP downstream effector pCREB in Nf1Prx1 muscles (Fig. 6A). The expression of multiple nuclear genes encoding mitochondrial proteins required for high-energy oxidative metabolism was also downregulated. Thus, a general bioenergetic crisis appears to be a common feature of various muscle dystrophies (19), including the one described here caused by the Nf1 deficiency.
Neurofibromin is required for the control of MAPK signaling in various cell types, including cardiac and muscle cells. MEK1 was previously shown to negatively regulate early steps of muscle differentiation (27). Additionally, constitutive activation of Ras and/or Raf kinase upstream of MEK1 was also reported to inhibit muscle cell differentiation (28,29). Importantly, MEK1 was shown to interact with the MyoD nuclear complex and inhibit its function (30). Thus, it appears likely that MEK1 hyperactivation contributes to the observed inhibition of muscle differentiation in Nf1Prx1 mice. In the current study, we detected increased levels of pMEK1 and pERK1/2 in the adult Nf1-deficient skeletal muscles as well as an increased number of pMEK1-positive cells in the muscle interfiber space of E16.5 Nf1-deficient muscles. However, we did not detect significant differences in the number of pMEK-positive myonuclei in E16.5 NF1-Prx1 mice, but at this stage we cannot rule out differences in signaling intensity. Further studies are needed to address the exact mechanism of the NF1 muscle dystrophy and to address which part of the muscular defect is cell-autonomous or non-cell-autonomous. Apart from activation of the canonical MAPK pathway components, we also detected hyperphosphorylation of CREB which is a known downstream target of PKA and RSK2 kinases (31). Both CREB1 and CREB2 (ATF4) were found to heterodimerize with MyoD and inhibit myogenic differentiation of 10T1/2 cells when overexpressed (32). Thus, the CREB factor is another candidate factor likely contributing to the muscle differentiation phenotype we observed.
Among several pathological changes, Nf1Prx1 mice invariably show cartilaginous fusions and/or partial fusions of the hip joints, a phenotype which is not observed in individuals with NF1. We have previously shown that this phenotype results from an early embryonic failure of joint cavitation (6 and Supplementary Material, Fig. S2). While joint fusions were consistently observed in Nf1Prx1 mice (Supplementary Material, Fig. S2), conditional inactivation of Nf1 in chondrocytes using the collagen type II alpha 1 (Col2a1) promoter did not result in such a phenotype (data not shown), suggesting that the cause of joint fusions in Nf1Prx1 mice was extra-cartilaginous. It has been shown that joint formation is critically dependent on embryonic movement, i.e. on the proper function of muscles. Reduction in movement either by denervation or by inhibition of muscle development results in joint fusions (33,34), indicating that reduced muscle strength as well as the absence of individual muscles may cause or at least contribute to the joint fusions in Nf1Prx1 mice. The skeletal muscles in Nf1Prx1 mice were significantly smaller in size and mutant mice displayed severely reduced muscle force in the force gauge pull test when compared with controls (Fig. 1C–G). It is important to mention that no differences in muscle strength or histological appearance were observed between WT and Nf1Prx1 heterozygous mice (data not shown), indicating that heterozygous gene inactivation was insufficient to cause overt muscle phenotype in the murine model. Strikingly, muscle groups involved in hip joint movement were most severely affected in homozygous Nf1Prx1 mice, with the gluteus maximus showing a 50% size reduction already at E13.5 (Fig. 5B) and severe reduction and fibrosis at postnatal day 2 (Supplementary Material, Fig. S2). Importantly, consistent with the causal relationship between the muscle formation defect and the joint fusion in Nf1Prx1 mice, the onset of the muscle defect preceded hip joint fusion (E12.5/13.5 versus E14.5). Similarly, we observed a coincidence of the spinal kyphosis in the Nf1Prx1 mice (Supplementary Material, Fig. S3) and the reduced size of m. latissimus dorsi primordia at E13.5 (Fig. 5B) as well as reduced thickness of m. latissimus dorsi in the postnatal stages (data not shown). Muscular dystrophy was previously implicated in pathogenesis of kyphosis in several mouse models including Mdx mice (35). Thus, it seems likely that the kyphosis in Nf1Prx1 mice is directly linked to the observed skeletal muscle dystrophy.
Altogether, the Nf1Prx1 phenotype described here clearly indicates that neurofibromin is required for skeletal muscle development and its absence results in muscle dystrophy. These findings highlight the importance of Ras signaling in development and maintenance of muscles and are of relevance for understanding the pathogenesis of NF1, in particular the skeletal system abnormalities such as scoliosis, osteoporosis and tibial dysplasia. Since the skeleton is directly influenced by muscle force, a reduction in muscle mass/force results in a reduction in bone mass and thus osteporosis. Likewise, a weakness of back musculature may contribute to the pathogenesis of scoliosis as seen in many muscular disorders, such as Duchenne muscular dystrophy.
MATERIALS AND METHODS
Mouse breeding and genotyping
Nf1flox and Rosa26-LacZ mice were bred and genotyped as described previously (36,37). See Supplementary material for primer sequences used for detection of the Cre transgene in the Prx1-Cre mouse strain. Recombination efficiency was tested in a competitive PCR using primers P1, P2 and P4 where P1 + P2 amplifies the excised allele (280 bp) and P1 + P4 amplifies the intact floxed allele (350 bp; for details see 32).
Protein and mRNA analysis
Whole triceps muscle lysates were prepared in radioimmunoprecipitation assay buffer buffer and protein concentration was determined with the bicinchoninic acid protein assay protein assay (Pierce). Twenty grams of protein were loaded per lane and resolved by electrophoresis in sodium dodecyl sulfate–polyacrylamide gels and transferred onto polyvinylidene fluoride membranes (Amersham). For western blot analysis, membranes were incubated with the following antibodies: anti-phospho-p42/44 (pERK1/2) #9102 (Cell Signaling), anti-p44 (ERK1) #4372 (Cell Signaling), anti-pCREB #4370S (Cell Signaling), anti-CREB #9197 (Cell Signaling), anti-pMEK1/2 #9121S (Cell Signaling) and anti-glycerinaldehyd-3-phosphat-dehydrogenase #Sc-25778 (Santa Cruz).
Histology and in situ hybridization
For histological analysis, adult limbs were fixed in 4% paraformaldehyd (PFA) containing 0.5% ethylene diamine tetraacetic acid (EDTA), dehydrated and embedded in paraffin. Heidenhain's Azan tricrome staining was performed according to the standard procedures.
For in situ hybridization, P22 limbs were fixed in 4% PFA/EDTA, dehydrated and embedded in paraffin. Sections were hybridized with digoxygenin-labeled mRNA probes as described previously (38). WISH was performed as described previously (39). Histomorphometric analysis was performed with AxioVision Outmess Software (Zeiss). See Supplementary material for primers used for generation of Collagen1a1-specific riboprobe template.
Immunohistology
E11.5, E14.5 or E16.5 embryos were fixed for 2 h in 4% PFA/phosphate buffer saline (PBS) at 4°C, washed several times in cold PBS and equilibrated in 20% sucrose/PBS at 4°C over night and embedded in compound for cryostat sectioning compound. Twelve micrometers of frozen sections were thawed for 4 h, washed in PBS and blocked in tyramide signal amplification (TSA) buffer (PerkinElmer Life Science) containing 10% Normal Horse Serum, in PBS. Antibodies against Lbx1 (kind gift from Dr Carmen Birchmeier) 1:200, MyoD #SC-304 (Santa Cruz) 1:100 and Desmin #SC-14026 (Santa Cruz) 1:200 were applied in 0.3% BSA; 0.3% Normal Horse Serum; 0.4% Triton X-100 in PBS and incubated over night at 4°C. Finally, sections were washed several times in PBST and incubated with Alexa-Fluor-conjugated secondary antibodies (Invitrogen). Sample examination was done with an AxioVert 200 fluorescence microscope and AxioVision software (both Zeiss).
Proliferation analysis was done on paraffin sections after in situ hybridization. Sections were treated for 30 min in 2 N HCl at 37°C, washed in PBS, blocked in 5% Normal Goat Serum, 3% BSA/PBS, incubated with anti-BrdU #1-299-964 antibody (Roche) 1:50.
pMEK1/2 #9121S (Cell Signaling) staining was done on paraffin sections using citrate buffer, permeabilization for 15 min in 5% Normal Goat Serum, 3% BSA, 0.1% Triton X-100 in TBS and blocking in 5% Normal Goat Serum, 3% BSA, 0.1% Tween-20/TBS solution. Anti-pMEK1/2 antibody was diluted 1:100, followed by goat anti-rabbit horseradish peroxidase (DAKOCytomation) incubation and TSA amplification (PerkinElmer Life Science) for 15 min at RT. Finally, Fitc-Streptavidin antibody (Invitrogen) was used. For control of antibody specificity, sections were pre-treated with alkaline phosphatse. No positive staining was observed on the dephosphorylated control sections (data not shown).
Satellite cell isolation
Satellite cells were isolated essentially as previously described (40), with following minor modifications. Bulk muscle tissue from muscle above the abdomen, including lattisimus and diaprhagm, was excised from adult mice and subjected to enzymatic digestion with dispase (BD) and collagenase (Worthington) for 1h. The resulting suspension was consecutively filtered over 100 and 40 µm cell strainers and subsequently subjected to fractionation using a discontinuous Percoll density gradient. Satellite cells were collected at the 70%/30% Percoll density interphase, washed and plated into collagen-coated plates for 12 h to allow fibroblasts to attach. Finally, the supernatant containing highly enriched satellite cells was recollected and plated into collagen-coated µclear 96-well plates (NUNC) and briefly centrifuged to facilitate satellite cell attachment.
EdU pulse proliferation assay. Satellite cells were pulsed with 10 µm EdU for 3 h, fixed and stained at the indicated time points. Cells were counted using ImageJ. After an initial growth phase of 3 days in rich media (Dulbecco's modified eagle's medium (DMEM) containing 30% fetal calf serum), satellite cells were subjected to differentiation media (DMEM containing 2% Horse Serum) and fixed at the indicated time points. Myotubes were labeled with antibody against meromyosin MF-20 (DSHB) 1:20.
Grip strength measurements
Grip force measurements were performed using a handheld Chatillon digital force gauge (Chatillon DFE Series Digital Force Gauge, Ametek Inc.) as previously described (41). Briefly, mice were held by the tail and gently lowered towards the apparatus. They were allowed to grip the grid with their forelimbs only and were pulled backwards in a horizontal plane. The maximum load measurement reflected the highest force applied to the grid, and the average load was the mean of the total strength. The mice were pre-conditioned before each measurement and the experiment was repeated two times per mouse.
Microarray hybridization and differential expression evaluation
RNA was isolated from the triceps muscles using peqGOLD TriFast (PeqLab Biotechnologie GmbH) according to the supplied protocol. The cRNA synthesis and microarray hybridizaton were done according to the standard protocols applied in the Laboratory of Functional Genome Research Charité, Core Facility. A total of eight high-density oligonucleotide mouse GENE 1.0 ST arrays (Affymetrix) were used in this study. Samples from each animal (four Nf1 and four WT) were applied individually to each array. All the procedures and hybridization were performed according to the Genechip expression technical manual (Affymetrix) as previously reported (42). Raw data were quantified with Microarray Suite (MAS 5.0) software. Expression data were normalized using the Genespring settings for Affymetrix gene chip arrays. Results from probe sets on each array were subjected to one-way ANOVA analysis to reveal the genes significantly deregulated in the Nf1-deficient muscles (F-factor ≥ 20). Array data have been made available through the GEO data repository of the National Centre for Biotechnology Information.
KEGG classification and the gene class enrichment analysis
We downloaded annotations including 6773 mouse genes and 207 pathways from the KEGG database (43). For each pathway, we calculated the number of overlaps between pathway genes and genes that were found to be significantly up- or downregulated based on the microarray data. We computed P-values for significant overlaps using Fisher's exact test with Bonferroni multiple-testing correction. Figure 3B shows the KEGG classes that we identified to be most significantly enriched in the set of upregulated genes. We detected no significantly enriched pathways in the set of downregulated genes.
Transcriptome comparison
We downloaded the Gene Expression Omnibus data set GSE7187 (18) and normalized raw intensity values using GC robust multi-array average background adjustment and identified differentially expressed genes with ANOVA and Benjamini–Hochberg multiple-testing correction (FDR < 0.05). For the human muscular dystrophies, we used the absolute intensities from Supplementary Material, Table S1 to define differential genes that showed at least 2-fold expression change in each comparison with the control samples (24). We restricted the comparison of differentially expressed genes to those that were probed on both MOE430A and MOUSE GENE 1.0 ST microarray platforms.
Statistical analysis
Experimental results are expressed as mean ± SEM. Numbers of animals used in the assays are indicated below graphs. Unless otherwise stated, differences between groups were evaluated using unpaired t-test with Welch's correction: *P < 0.05; **P < 0.01; ***P < 0.001.
SUPPLEMENTARY MATERIAL
FUNDING
M.K. and N.K. were supported by the Young Investigator Award from Children Tumour Fundation, New York, Grant 2007-01-038 and by Bundesministerium für Bildung und Forschung (BMBF), Grant NF1-01GM0844 (M.K., S.M.). D.A.S. was supported by grant K23 NS052500 from the National Institute of Neurological Disorders and Stroke, a Doris Duke Charitable Foundation Clinical Scientist Development Award, and the Thrasher Research Fund. J.K. and T.B. were supported by the Excellence Cluster Cardio Pulmonary System (ECCPS). This work was also supported by the Sixth Framework of the European Commission, EuroGrow project LSHM-CT-2007-037471, and by grant from Berlin-Brandenburg Center for Regenerative Therapies (Platform A nr-30).
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank Dr Ute Ungethuem for performing microarray hybridizations and Ralf J. Kuban for primary evaluation of the microarray hybridizations data and differential gene expression analysis. We are also thankful to Dr Carmen Birchmeier for sharing her expertise with us.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stevenson D.A., Zhou H., Ashrafi S., Messiaen L.M., Carey J.C., D'Astous J.L., Santora S.D., Viskochil D.H. Double inactivation of NF1 in tibial pseudarthrosis. Am. J. Hum. Genet. 2006;79:143–148. doi: 10.1086/504441. doi:10.1086/504441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiest V., Eisenbarth I., Schmegner C., Krone W., Assum G. Somatic NF1 mutation spectra in a family with neurofibromatosis type 1: toward a theory of genetic modifiers. Hum. Mutat. 2003;22:423–427. doi: 10.1002/humu.10272. doi:10.1002/humu.10272. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y.Y., Vik T.A., Ryder J.W., Srour E.F., Jacks T., Shannon K., Clapp D.W. Nf1 regulates hematopoietic progenitor cell growth and ras signaling in response to multiple cytokines. J. Exp. Med. 1998;187:1893–1902. doi: 10.1084/jem.187.11.1893. doi:10.1084/jem.187.11.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegedus B., Dasgupta B., Shin J.E., Emnett R.J., Hart-Mahon E.K., Elghazi L., Bernal-Mizrachi E., Gutmann D.H. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1:443–457. doi: 10.1016/j.stem.2007.07.008. doi:10.1016/j.stem.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Wu X., Estwick S.A., Chen S., Yu M., Ming W., Nebesio T.D., Li Y., Yuan J., Kapur R., Ingram D., et al. Neurofibromin plays a critical role in modulating osteoblast differentiation of mesenchymal stem/progenitor cells. Hum. Mol. Genet. 2006;15:2837–2845. doi: 10.1093/hmg/ddl208. doi:10.1093/hmg/ddl208. [DOI] [PubMed] [Google Scholar]
- 6.Kolanczyk M., Kossler N., Kuhnisch J., Lavitas L., Stricker S., Wilkening U., Manjubala I., Fratzl P., Sporle R., Herrmann B.G., et al. Multiple roles for neurofibromin in skeletal development and growth. Hum. Mol. Genet. 2007;16:874–886. doi: 10.1093/hmg/ddm032. doi:10.1093/hmg/ddm032. [DOI] [PubMed] [Google Scholar]
- 7.Elefteriou F., Benson M.D., Sowa H., Starbuck M., Liu X., Ron D., Parada L.F., Karsenty G. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441–451. doi: 10.1016/j.cmet.2006.10.010. doi:10.1016/j.cmet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elefteriou F., Kolanczyk M., Schindeler A., Viskochil D.H., Hock J.M., Schorry E.K., Crawford A.H., Friedman J.M., Little D., Peltonen J., et al. Skeletal abnormalities in neurofibromatosis type 1: approaches to therapeutic options. Am. J. Med. Genet. A. 2009;149A:2327–2338. doi: 10.1002/ajmg.a.33045. doi:10.1002/ajmg.a.33045. [DOI] [PubMed] [Google Scholar]
- 9.Souza J.F., Passos R.L., Guedes A.C., Rezende N.A., Rodrigues L.O. Muscular force is reduced in neurofibromatosis type 1. J. Musculoskelet. Neuronal Interact. 2009;9:15–17. [PubMed] [Google Scholar]
- 10.Stevenson D.A., Moyer-Mileur L.J., Carey J.C., Quick J.L., Hoff C.J., Viskochil D.H. Case-control study of the muscular compartments and osseous strength in neurofibromatosis type 1 using peripheral quantitative computed tomography. J. Musculoskelet. Neuronal Interact. 2005;5:145–149. [PubMed] [Google Scholar]
- 11.Brohmann H., Jagla K., Birchmeier C. The role of Lbx1 in migration of muscle precursor cells. Development. 2000;127:437–445. doi: 10.1242/dev.127.2.437. [DOI] [PubMed] [Google Scholar]
- 12.Schafer K., Braun T. Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat. Genet. 1999;23:213–216. doi: 10.1038/13843. doi:10.1038/13843. [DOI] [PubMed] [Google Scholar]
- 13.Birchmeier C., Brohmann H. Genes that control the development of migrating muscle precursor cells. Curr. Opin. Cell Biol. 2000;12:725–730. doi: 10.1016/s0955-0674(00)00159-9. doi:10.1016/S0955-0674(00)00159-9. [DOI] [PubMed] [Google Scholar]
- 14.Chevallier A., Kieny M. On the role of the connective tissue in the patterning of the chick limb musculature. Wilhelm Roux Arch. 1982;191:277–280. doi: 10.1007/BF00848416. doi:10.1007/BF00848416. [DOI] [PubMed] [Google Scholar]
- 15.Hasson P., DeLaurier A., Bennett M., Grigorieva E., Naiche L.A., Papaioannou V.E., Mohun T.J., Logan M.P. Tbx4 and tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev. Cell. 2010;18:148–156. doi: 10.1016/j.devcel.2009.11.013. doi:10.1016/j.devcel.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kardon G., Campbell J.K., Tabin C.J. Local extrinsic signals determine muscle and endothelial cell fate and patterning in the vertebrate limb. Dev. Cell. 2002;3:533–545. doi: 10.1016/s1534-5807(02)00291-5. doi:10.1016/S1534-5807(02)00291-5. [DOI] [PubMed] [Google Scholar]
- 17.Francis-West P.H., Antoni L., Anakwe K. Regulation of myogenic differentiation in the developing limb bud. J. Anat. 2003;202:69–81. doi: 10.1046/j.1469-7580.2003.00136.x. doi:10.1046/j.1469-7580.2003.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baban D., Davies K.E. Microarray analysis of mdx mice expressing high levels of utrophin: therapeutic implications for dystrophin deficiency. Neuromuscul. Disord. 2008;18:239–247. doi: 10.1016/j.nmd.2007.11.011. doi:10.1016/j.nmd.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Lecker S.H., Jagoe R.T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S.R., Mitch W.E., Goldberg A.L. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. doi:10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 20.Bione S., D'Adamo P., Maestrini E., Gedeon A.K., Bolhuis P.A., Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat. Genet. 1996;12:385–389. doi: 10.1038/ng0496-385. doi:10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 21.Marti A., Larrarte E., Novo F.J., Garcia M., Martinez J.A. UCP2 muscle gene transfer modifies mitochondrial membrane potential. Int. J. Obes. Relat. Metab. Disord. 2001;25:68–74. doi: 10.1038/sj.ijo.0801484. doi:10.1038/sj.ijo.0801484. [DOI] [PubMed] [Google Scholar]
- 22.Gredinger E., Gerber A.N., Tamir Y., Tapscott S.J., Bengal E. Mitogen-activated protein kinase pathway is involved in the differentiation of muscle cells. J. Biol. Chem. 1998;273:10436–10444. doi: 10.1074/jbc.273.17.10436. doi:10.1074/jbc.273.17.10436. [DOI] [PubMed] [Google Scholar]
- 23.Xu J., Ismat F.A., Wang T., Lu M.M., Antonucci N., Epstein J.A. Cardiomyocyte-specific loss of neurofibromin promotes cardiac hypertrophy and dysfunction. Circ. Res. 2009;105:304–311. doi: 10.1161/CIRCRESAHA.109.201509. doi:10.1161/CIRCRESAHA.109.201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y.W., Zhao P., Borup R., Hoffman E.P. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J. Cell Biol. 2000;151:1321–1336. doi: 10.1083/jcb.151.6.1321. doi:10.1083/jcb.151.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Blagden C., Fan J., Nowak S.J., Taniuchi I., Littman D.R., Burden S.J. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005;19:1715–1722. doi: 10.1101/gad.1318305. doi:10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter R.B., Kandarian S.C. Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J. Clin. Invest. 2004;114:1504–1511. doi: 10.1172/JCI21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jo C., Jang B.G., Jo S.A. MEK1 plays contrary stage-specific roles in skeletal myogenic differentiation. Cell Signal. 2009;21:1910–1917. doi: 10.1016/j.cellsig.2009.08.008. doi:10.1016/j.cellsig.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Ramocki M.B., Johnson S.E., White M.A., Ashendel C.L., Konieczny S.F., Taparowsky E.J. Signaling through mitogen-activated protein kinase and Rac/Rho does not duplicate the effects of activated Ras on skeletal myogenesis. Mol. Cell Biol. 1997;17:3547–3555. doi: 10.1128/mcb.17.7.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page J.L., Wang X., Sordillo L.M., Johnson S.E. MEKK1 signaling through p38 leads to transcriptional inactivation of E47 and repression of skeletal myogenesis. J. Biol. Chem. 2004;279:30966–30972. doi: 10.1074/jbc.M402224200. doi:10.1074/jbc.M402224200. [DOI] [PubMed] [Google Scholar]
- 30.Perry R.L., Parker M.H., Rudnicki M.A. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol. Cell. 2001;8:291–301. doi: 10.1016/s1097-2765(01)00302-1. doi:10.1016/S1097-2765(01)00302-1. [DOI] [PubMed] [Google Scholar]
- 31.Xing J., Ginty D.D., Greenberg M.E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. doi:10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 32.Muir T., Wilson-Rawls J., Stevens J.D., Rawls A., Schweitzer R., Kang C., Skinner M.K. Integration of CREB and bHLH transcriptional signaling pathways through direct heterodimerization of the proteins: role in muscle and testis development. Mol. Reprod. Dev. 2008;75:1637–1652. doi: 10.1002/mrd.20902. doi:10.1002/mrd.20902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rot-Nikcevic I., Reddy T., Downing K.J., Belliveau A.C., Hallgrimsson B., Hall B.K., Kablar B. Myf5-/- :MyoD-/- amyogenic fetuses reveal the importance of early contraction and static loading by striated muscle in mouse skeletogenesis. Dev. Genes Evol. 2006;216:1–9. doi: 10.1007/s00427-005-0024-9. doi:10.1007/s00427-005-0024-9. [DOI] [PubMed] [Google Scholar]
- 34.Kahn J., Shwartz Y., Blitz E., Krief S., Sharir A., Breitel D.A., Rattenbach R., Relaix F., Maire P., Rountree R.B., et al. Muscle contraction is necessary to maintain joint progenitor cell fate. Dev. Cell. 2009;16:734–743. doi: 10.1016/j.devcel.2009.04.013. doi:10.1016/j.devcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Laws N., Hoey A. Progression of kyphosis in mdx mice. J. Appl. Physiol. 2004;97:1970–1977. doi: 10.1152/japplphysiol.01357.2003. doi:10.1152/japplphysiol.01357.2003. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y., Romero M.I., Ghosh P., Ye Z., Charnay P., Rushing E.J., Marth J.D., Parada L.F. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. doi:10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolanczyk M., Kuhnisch J., Kossler N., Osswald M., Stumpp S., Thurisch B., Kornak U., Mundlos S. Modelling neurofibromatosis type 1 tibial dysplasia and its treatment with lovastatin. BMC Med. 2008;6:21. doi: 10.1186/1741-7015-6-21. doi:10.1186/1741-7015-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stricker S., Fundele R., Vortkamp A., Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev. Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. doi:10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 39.Witte F., Dokas J., Neuendorf F., Mundlos S., Stricker S. Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Expr. Patterns. 2009;9:215–223. doi: 10.1016/j.gep.2008.12.009. doi:10.1016/j.gep.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Yablonka-Reuveni Z., Nameroff M. Skeletal muscle cell populations. Separation and partial characterization of fibroblast-like cells from embryonic tissue using density centrifugation. Histochemistry. 1987;87:27–38. doi: 10.1007/BF00518721. doi:10.1007/BF00518721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pirog K.A., Jaka O., Katakura Y., Meadows R.S., Kadler K.E., Boot-Handford R.P., Briggs M.D. A mouse model offers novel insights into the myopathy and tendinopathy often associated with pseudoachondroplasia and multiple epiphyseal dysplasia. Hum. Mol. Genet. 2010;19:52–64. doi: 10.1093/hmg/ddp466. doi:10.1093/hmg/ddp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross J., Machulik A., Amarjargal N., Moller R., Ungethum U., Kuban R.J., Fuchs F.U., Andreeva N., Fuchs J., Henke W., et al. Expression of apoptosis-related genes in the organ of Corti, modiolus and stria vascularis of newborn rats. Brain Res. 2007;1162:56–68. doi: 10.1016/j.brainres.2007.05.061. doi:10.1016/j.brainres.2007.05.061. [DOI] [PubMed] [Google Scholar]
- 43.Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. doi:10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.