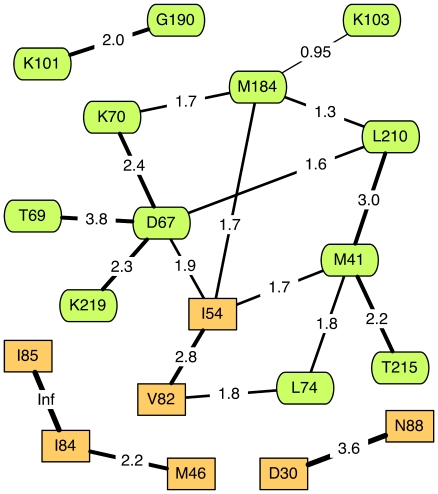
Figure 2. Bayesian network depicting conditional dependencies among surveillance drug resistance mutations (SDRMs) in ARV-naïve sequences.
Each labeled node corresponds to a position in protease [rectangular] or RT (rounded). Nodes without dependencies (L23, L24, V32, I47, I50, F53, G73, L76, K65, V75, F77, Y115, F116, L100, V106, V179, Y181, and Y188) were omitted from the graph for clarity. Connections between nodes (edges) are labeled with the log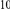 odds ratio of the
odds ratio of the 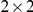 contingency table for the presence/absence of SDRMs at the respective sites, e.g., an SDRM at position PR-D30 is estimated to be
contingency table for the presence/absence of SDRMs at the respective sites, e.g., an SDRM at position PR-D30 is estimated to be 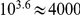 times more likely to be present in an ARV-naïve sequence that contains an SDRM at position PR-N88. Line widths for edges are also drawn in proportion to the log
times more likely to be present in an ARV-naïve sequence that contains an SDRM at position PR-N88. Line widths for edges are also drawn in proportion to the log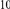 odds ratios. Inf = infinity, i.e., unable to estimate because of zero count(s) in the contingency table.
odds ratios. Inf = infinity, i.e., unable to estimate because of zero count(s) in the contingency table.