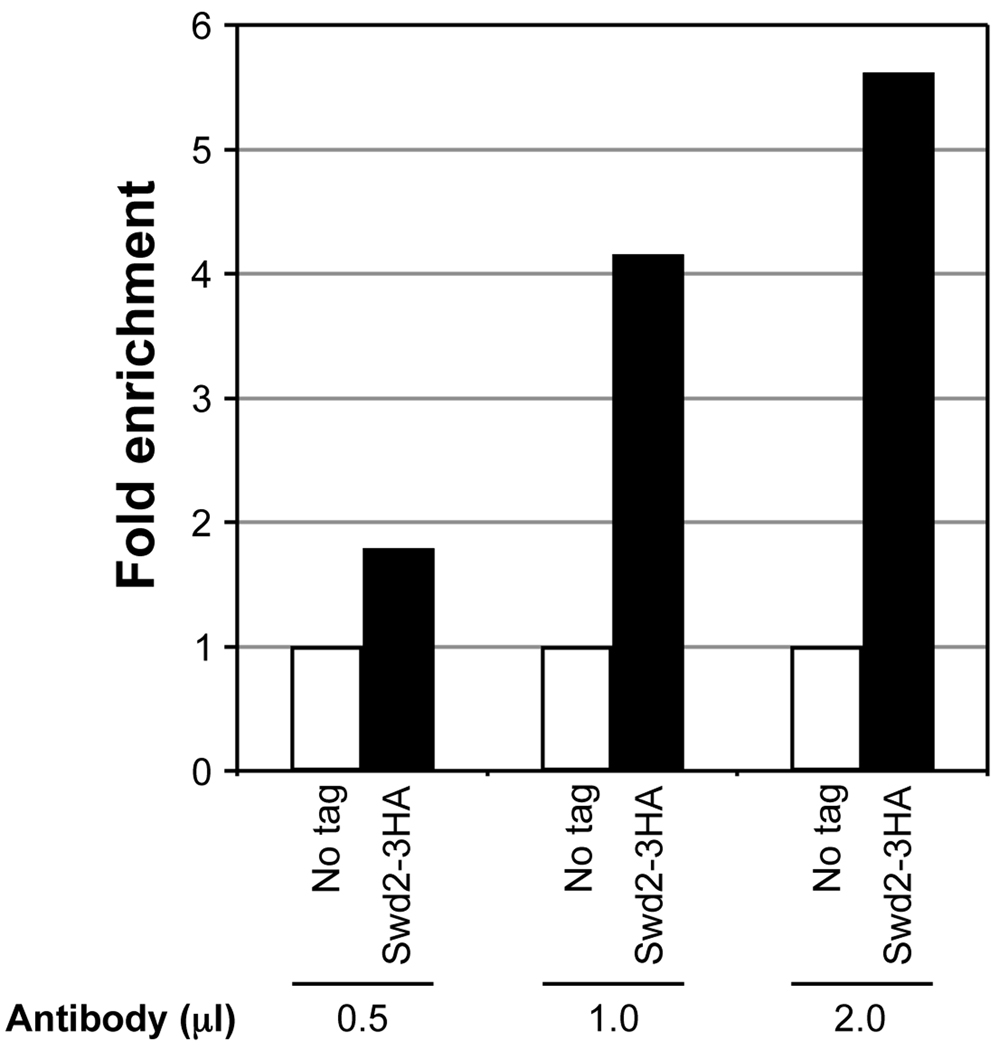
Fig. 2.
Antibody titration to determine the optimal amount required for high enrichment in ChIP assay. Soluble chromatin was prepared using a yeast strain expressing Swd2 with 3 copies of the HA epitope tag at the C-terminus (Swd2-3HA) or lacking the tag (no tag) subjected to double crosslinking (DMA and HCHO). The soluble chromatin (500 µg) was subjected to immunoprecipitation (IP) using 0.5 µl, 1 µl or 2 µl of an antibody that recognizes the HA epitope (α-HA, GenScript, catalog no. A00168). Quantitative PCR in real time (qPCR) was used to measure DNA associated with Swd2 in the immunoprecipitate and the total DNA in the soluble chromatin subjected to IP (input). Relative enrichment (IP/input) was initially calculated. For no tag control, the relative enrichment obtained represents the background signal (set as 1). Fold enrichment relative to the no tag control is shown for Swd2-3HA using different amounts of the antibody.