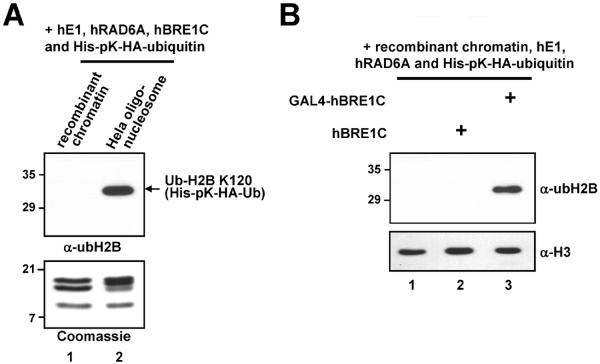
Fig. 4.
Altered chromatin structure and/or human BRE1 complex recruitment to chromatin is crucial for efficient H2B ubiquitylation. (A) The chromatin ubiquitylation assay was performed with the indicated substrates for H2B ubiquitylation (top). Analyses of purified recombinant histone octamers and natural oligonucleosome derived from HeLa cells by SDS-PAGE with Coomassie Blue staining to ensure comparable levels of protein usage for in vitro chromatin ubiquitylation assay (bottom). Note that endogenous ubH2B is not detected in lane 2 because a much smaller amount (350 ng) of nucleosome substrate was used in this assay. (B) hBRE1C recruitment to chromatin is critical for efficient H2B ubiquitylation. A recombinant chromatin template containing the pG5ML plasmid was subjected to the in vitro ubiquitylation assay with hE1, hRAD6A, ubiquitin and either hBRE1C or GAL4-hBRE1C, as indicated, and in the continued presence of the chromatin assembly factors. Reaction products were analyzed by immunoblot with indicated antibodies.