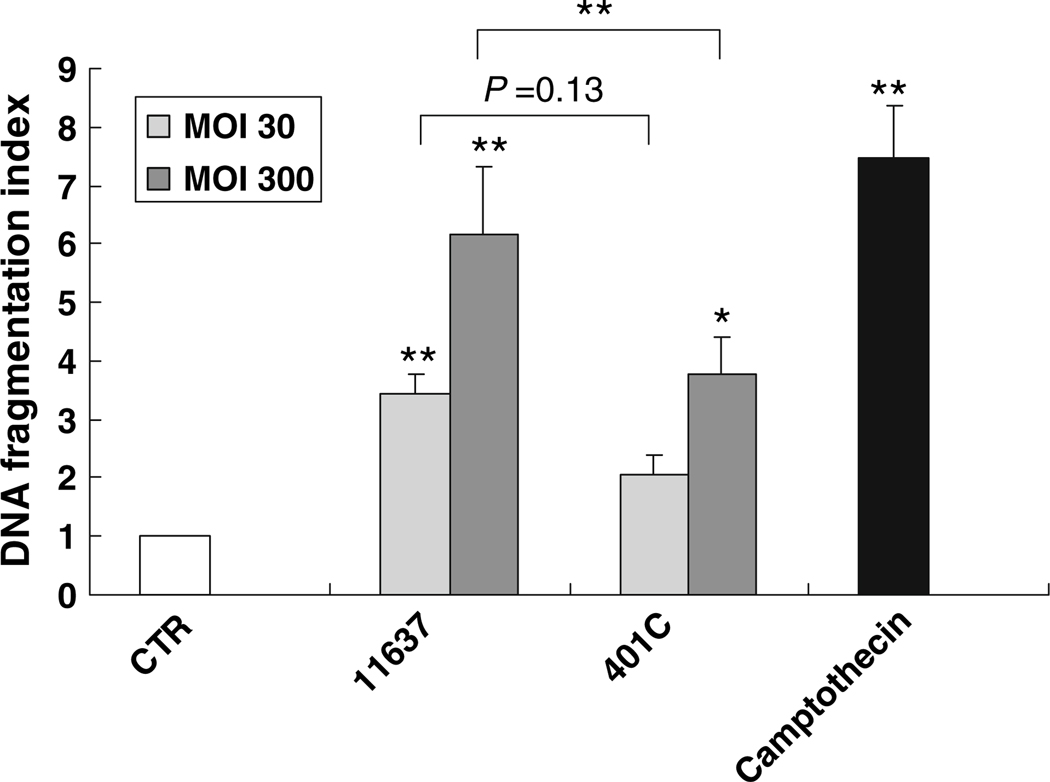
Fig. 3.
Comparison of DNA fragmentation in Huh7 cells co-incubated with or without H. pylori 11637 or 401C by quantitative apoptosis ELISA. Quantitative apoptosis ELISA methods quantify histon-complexed DNA fragments, which are biological hallmarks of apoptosis. DNA fragmentation index (405 nm absorbance of samples per 405 nm absorbance of control) was used for comparison of DNA fragmentation quantity. Huh7 cells were co-cultured with H. pylori 11637 or 401C at MOI of 30 (light gray bars) or 300 (dark gray bars) for 48 h. Control cells were incubated with cell culture medium absent with H. pylori (white bar). 2 µg/ml of camptothecin was used for positive control (black bar). Data are presented as mean ± SEM. **P ≤ 0.01, *P < 0.05