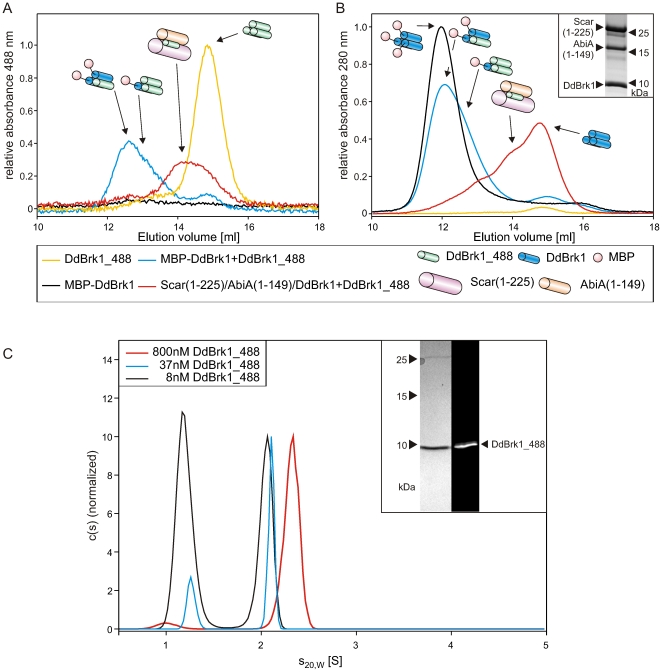
Figure 5. DdBrk1 chains swap within DdBrk1-containing complexes.
MBP-DdBrk1 homotrimers and Scar(1-225)/AbiA(1-149)/DdBrk1-complexes were incubated with DdBrk1_488 and subjected to gelfiltration. (A) The elution of 200 nM DdBrk1_488 alone and 100 nM DdBrk1_488 after incubation with other DdBrk1 containing complexes was monitored at 488 nm. 100 nM DdBrk1_488 showed an increased rH in the presence of 1.4 µM MBP-DdBrk1- and Scar(1–225)/AbiA(1–149)/DdBrk1-complexes, confirming an exchange of subunits within these complexes. The unlabeled complexes were undetectable in the absence of DdBrk1_488 at 488 nm absorption. (B) Simultaneously recorded elution profiles at 280 nm showed slightly decreased hydrodynamic radii of the MBP-DdBrk1 fusion construct after incubation with DdBrk1_488, indicating different stoichiometries of trimeric MBP-DdBrk1/DdBrk1_488-complexes. The inset shows a 16% Tris-Tricine SDS-PAGE stained with Coomassie blue of copurified Scar(1–225)/AbiA(1–149)/DdBrk1 used in these experiments. For reasons of clarity, the different complexes are schematically shown in addition to the elution profiles. (C) Sedimentation velocity experiments of DdBrk1_488 showed dissociation of the homotrimer in the lower nanomolar range. The decreasing s-value of the faster sedimenting boundary at decreasing protein concentrations indicates a fast dissociation of the homotrimer when compared to the time scale of the experiment. The inset shows a 16% Tris-Tricine SDS-PAGE of DdBrk1_488 visualized by Coomassie-blue stain and ultraviolet illumination.