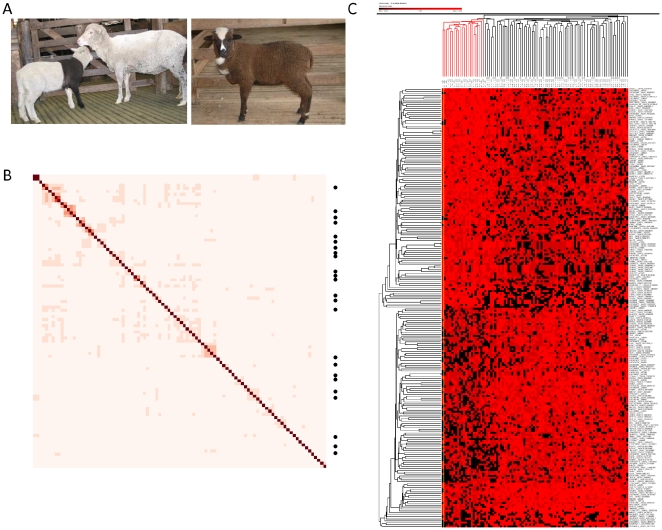
Figure 1. Genome wide association for piebald.
(A) A piebald lamb is pictured with its non-pigmented mother (left hand side). The asymmetrical presentation of pigmentation (the lamb has one pigmented and one white forelimb) characterises this colour morph from Recessive Black (right hand side) which arises through action of the Agouti locus [39]. (B) The genetic relationship between 24 piebald animals and 72 non-pigmented controls. Allele sharing, or genetic similarity, was calculated between each pair-wise combination of animal using 48,686 SNP. Increasing values of allele sharing are represented using darker colour. Unsupervised hierarchical clustering distributed the piebald cases (indicated with a dot) across the matrix, reflecting the sampling strategy used to select controls which were as closely related to cases as possible. (C) Unsupervised hierarchical clustering of the 226 associated SNP (P<0.001) successfully distinguished all piebald animals as genetically distinct from the controls. Animals are arranged into columns and annotated above the matrix using either red (piebald) or black lines (controls). The 226 associated SNP are arranged into rows and each cell indicates the observed genotypic outcome as follows: homozygote (bright red), heterozygote (dark red), alternate homozygote (black).