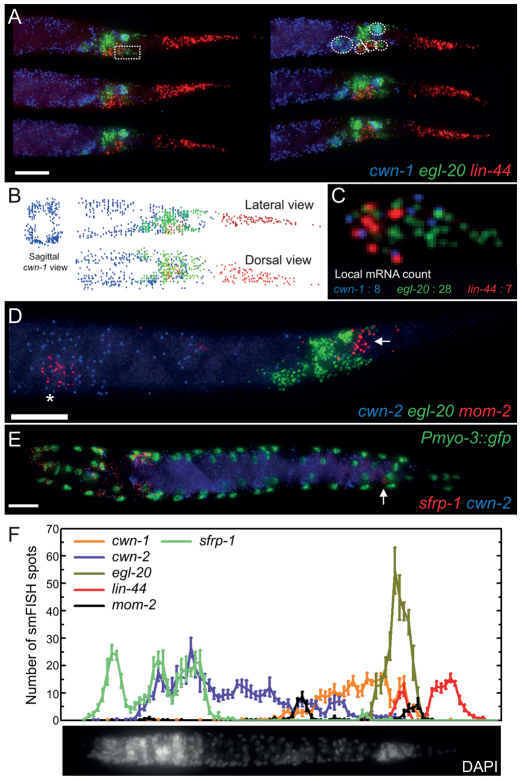
Fig. 1.
Single molecule mRNA FISH analyses of the C. elegans Wnt genes and sfrp-1 in staged L1 animals. Images are maximum intensity projections of lateral z-stacks. (A) Detection of cwn-1, egl-20 and lin-44 transcripts in the L1 larval tail of six individual animals. Broken circles indicate: top, the anal depressor muscle; from bottom left to right, P11/P12, Y, B and the body wall muscle cells VL23/VL24. The rectum is indicated by a solid line. Scale bar: 10 μm. (B) Transcript identification using a custom program written in MATLAB. A sagittal view of cwn-1 smFISH spots shows predominant expression in the four body wall muscle quadrants. (C) Magnification of the boxed area in A. smFISH spot counts of cwn-1, egl-20 and mom-2 are indicated. (D) Expression of cwn-2, egl-20 and mom-2 in the posterior half of the animal. The asterisk indicates the Z2 and Z3 germ line precursor cells, the arrow indicates the position of the tail cells that transiently express mom-2. Scale bar: 10 μm. (E) Expression of sfrp-1 and cwn-2 in an L1 larva. The nuclei of body wall muscle cells are highlighted by nuclear GFP. The posterior ventral nerve cord neuron expressing sfrp-1 was identified as DA7 (indicated by an arrow). Scale bar: 10 μm. (F) Quantification of Wnt and sfrp-1 smFISH spots along the anteroposterior axis of early L1 stage larvae. A DAPI stained animal is included for orientation. For all images, anterior is towards the left and posterior is towards the right. Error bars indicate s.e.m. (cwn-1, n=10; cwn-2, egl-20 and lin-44, n=13; mom-2, n=6 and sfrp-1, n=9).