Abstract
Peripheral isolated populations may undergo rapid divergence from the main population due to various factors such as a bottleneck or a founder effect followed by genetic drift or local selection pressures. Recent populations of two economically important Copadichromis species in Lake Malombe, a satellite lake of Lake Malawi, were neither genetically nor morphometrically distinct from their source populations in the main lake. Evidence was found for a founder effect which had a different impact on the genetic composition of the two species. In addition, the increased fishing pressure in Lake Malombe may have led to a reduction of the body sizes of both species.
1. Introduction
Peripheral populations are isolated or semi-isolated from the major distribution range of a species. The effects of geographical or ecological isolation and distinct environmental conditions can promote peripheral populations to diverge both genetically and phenotypically from core populations as a result of genetic drift and natural selection [1]. Peripheral populations are often smaller than core populations which can predestine such populations to bottleneck events [2]. This leads to the prediction that neutral genetic variations will be depleted in these populations. Nonetheless, peripheral populations may significantly contribute to a species' evolutionary potential by providing a source of adaptive genetic variance in a particular environmental setting [3, 4].
Taxa with island-like distribution patterns, such as the endemic cichlid fishes of the East African Great Lakes, provide an excellent opportunity for the study of evolutionary processes. Although many studies on these species have focussed on mechanisms of intralacustrine diversification within each of the main lakes [5, 6], the occurrence of endemic cichlids in Lake Nabugabo, a satellite lake isolated from Lake Victoria, initially raised the awareness of the evolutionary potential of peripheral isolates in the Great Lake region [7]. However, it was only recently that evidence was provided for the evolution of a cichlid species in an isolated satellite lake of Lake Malawi [8]. A genetically and phenotypically unique population of Rhamphochromis, a genus previously thought to be restricted to Lake Malawi and its permanently connected waters, was detected in Lake Chilingali. These examples suggest a role for (periodic) peripheral isolates in the evolutionary diversification of East African cichlids. In the present study, we evaluated empirical data on two economically important cichlid species to test whether peripheral populations from Lake Malombe, a satellite lake of Lake Malawi, have developed different genetic and phenotypic properties after recent colonisation from Lake Malawi.
Lake Malombe is a shallow, turbid, and nutrient-rich lake of about 390 km² with a maximum depth of only 5–7 m. The lake was formed by the recent (1930s) inundation of a flood plain of the Upper Shire River, about 15 km from its source in Lake Malawi [9]. Its ichthyofauna dominated by haplochromine cichlids is similar to that of Lake Malawi, but its diversity is lower and its relative productivity is higher. The lake supports an intensive fishery providing nearly 10% (ca. 4,000 t.y−1) of the total yield (ca. 50–60,000 t.y−1) within the Lake Malawi catchment [10, 11]. Since the mid-1980s, fishing effort on Lake Malombe has increased rapidly and mesh sizes have progressively been reduced to 19 mm or even less. Lake Malombe's haplochromine fishery yielded 9,500 tons in 1990 but this figure has declined to less than 4,000 tons in 2001 [12]. Among the economically most important haplochromines, Copadichromis sp. “Virginalis kajose” (cited as C. virginalis) landings declined by more than 50%, from 937 t.y−1 in 1991 to 412 t.y−1in 2001, while in contrast, C. chrysonotus catches increased from about 120 t.y−1 to 510 t.y−1 over the same time period with a similar fishing effort [12].
Their semi-isolated state, the distinct habitat conditions and the elevated fishing pressure, may have caused the Lake Malombe populations of Copadichromis sp. “Virginalis kajose” and C. chrysonotus to be susceptible to genetic and phenotypic divergence from their conspecifics in the main lake. It has been shown that the genetic variability of a population can be substantially reduced as a result of a founder effect following colonization and/or elevated fishing mortality [13, 14]. To test for a depletion of the genetic variability in the Lake Malombe populations, we analysed mitochondrial DNA (mtDNA) sequence data and microsatellite DNA variation. MtDNA variation is particularly sensitive to bottlenecks and random fluctuations in genotype frequencies, as its effective population size is one-fourth of nuclear alleles [15]. Microsatellites may be particularly informative in detecting structure among recently colonized or geographically proximal populations because of the much higher mutation rate than for mtDNA [16]. Furthermore, we looked for morphometric differences between populations from lakes Malombe and Malawi in response to the distinct habitat conditions. Haplochromine cichlids have been shown to be morphologically exceedingly versatile in response to environmental factors [17–19]. We expected morphometric differences between populations from Lake Malombe and Lake Malawi to be higher than among populations within Lake Malawi.
2. Materials and Methods
2.1. Sample Collection
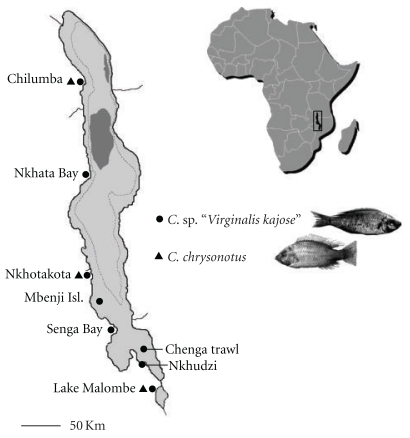
A total of 427 C. sp. “Virginalis kajose” and 192 C. chrysonotus specimens were sampled from one site in Lake Malombe and, respectively, seven and two localities in Lake Malawi (Figure 1). All fishes were collected with open water seines (chirimila nets), purse seines (nkacha nets), and beach seines by artisanal fishermen, except for one sample of C. sp. “Virginalis kajose” that was caught by a commercial trawler. Pelvic fin clips were preserved in absolute ethanol and stored at room temperature. The specimens were fixed in 10% formalin and are curated at the Royal Museum for Central Africa in Tervuren, Belgium. All available samples were used in the analysis of microsatellite DNA variability, except for C. chrysonotus from Senga Bay because of its low sample size. A subset of ten to twelve samples per species per locality were sequenced for an analysis of mtDNA haplotype variation, except for C. sp. “Virginalis kajose” from Lake Malombe of which a total of 24 specimens were sequenced. For the morphological analysis, we examined approximately 20 specimens per species from each locality.
Figure 1.
Map of Lakes Malawi and Malombe, East Africa. Sampling localities for Copadichromis sp. “Virginalis kajose” and C. chrysonotus are shown. Dashed bathymetric line indicates zones deeper than 250 metres; dark grey areas indicate lake zones deeper than 500 metres.
2.2. DNA Preparation and Amplification
Total genomic DNA was extracted using proteinase K digestion and salt precipitation, according to Aljanabi and Martinez [20]. DNA extracts were resuspended in 100 μL of autoclaved ddH2O. All samples were examined for genetic variation at 6 microsatellite markers: Pzeb1, Pzeb3, Pzeb4, Pzeb5 [21], UNH002 [22], and TmoM11 [23]. PCRs were performed under the following conditions: 94°C for 120 s, followed by 5 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 45 s, and followed by 30 cycles of 90°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by 72°C for 10 min. 10 μL reaction cocktails included 1 μL template DNA, 0.5 μM of each primer, 200 μM of each dNTP, 0.26 units Taq polymerase (Sigma Aldrich, Germany), and 1 μL 10x reaction buffer (Sigma Aldrich). PCR amplification products were run on 6% denaturing polyacrylamide gels using an ALF Express DNA Sequencer (Amersham Pharmacia Biotech). Fragment sizes were scored with ALFWin Fragment Analyser v1.0 (Amersham Pharmacia Biotech), using M13mp8 DNA standards as external references, following van Oppen et al. [21].
Mitochondrial DNA sequence variation was examined in a 325 bp fragment of the control region (D-loop). PCR amplification was carried out in a 25 μL buffered reaction mixture, containing 5 μL template DNA, 0.5 μM of each primer, H16498 and L15995 [24], 200 μM of each dNTP, 2.5 μL of 10x buffer (1 mM MgCl2), and 0.65 units of Red Taq Polymerase (Sigma Aldrich). PCR conditions consisted of an initial denaturation at 94°C for 120 s, followed by 35 cycles of 94°C for 60 s, 52°C for 60 s, and 72°C for 120 s, followed by 72°C for 10 min. The success of amplification was checked by electrophoresis on 1% agarose gel. The PCR products were purified following the qiaquick PCR purification kit protocol, prior to being added as template for chain-termination sequencing, carried out in 10 μL reaction volumes, using 2 μL purified DNA template, 1 μL 5x Sequencing Buffer (Applied Biosystems), 2 μL Ready Reaction Mix (Applied Biosystems), and 2 μL primer (L15995, 2 μM). The sequencing program consisted of an initial denaturation step at 96°C for 60 s, followed by 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 240 s. DNA fragments were purified by ethanol precipitation and subsequently visualized on a 3130 capillary sequencer (Applied Biosystems).
2.3. Analysis of DNA Variation
Nuclear genetic diversity estimates, such as allele numbers and observed and unbiased expected number of heterozygotes at each locus, were calculated using GENETIX 4.02 [25]. Departures from Hardy-Weinberg equilibrium and linkage disequilibrium were tested using exact tests as implemented by GENEPOP 3.3 [26]. Significance levels were obtained by the Markov chain method using 5,000 dememorisations, 500 batches, and 2,000 iterations per batch. Allelic richness (AR) and estimates of global population differentiation using Wright's FST [27] and pairwise FST values, estimated by θ [28], were calculated in FSTAT 2.9.3 [29]. Significance of FST values was calculated in Genepop by the Markov chain method using 5,000 dememorisations, 2,000 permutations, and 500 batches in an exact test for population differentiation. Recent population size reduction was examined by testing for heterozygosity excess and mode shift in allele frequency with the program BOTTLENECK 1.2.02 [30]. Following the recommendations by Luikart and Cornuet [31] for microsatellite data, we used the Wilcoxon signed-rank test with a two-phased model of mutation (TPM). The parameters chosen for TPM were: variance 30% and probability for SMM 70%, and estimations were based on 1000 iterations.
After manual correction in Chromas 1.45, the mtDNA sequences were aligned using ClustalW [32] and visually checked afterwards. Haplotype frequencies, number of segregating sites, mean number of pairwise differences, gene diversity (H), and nucleotide diversity (π) were calculated with DNAsp [33]. For the two fish species, a mtDNA haplotype network was constructed with the program TCS (v. 1.21) using statistical parsimony [34].
2.4. Analysis of Morphometric Variation
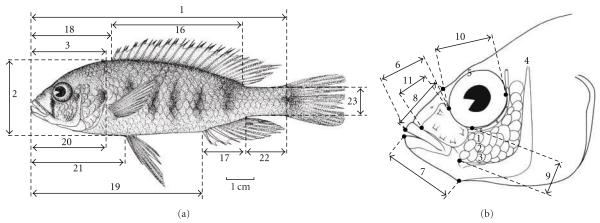
We measured 19 metric characters (Figure 2) as defined by Snoeks [35]. The measurements were taken with dial callipers under a binocular microscope. The 19 measurements were standard length (SL), body depth (BD), head length (HL), head width (HW), interorbital width (IOW), snout length (SnL), lower jaw length (LJL), premaxillary pedicel length (PPL), cheek depth (ChD), eye diameter (ED), lacrimal depth (LaD), dorsal fin base length (DFB), anal fin base length (AFB), predorsal fin distance (PRD), preanal fin distance (PRA), prepectoral fin distance (PRP), preventral fin distance (PRV), caudal peduncle length (CPL) and caudal peduncle depth (CPD). These measurements are typically used in taxonomic research on cichlids, where they have proven to be useful to detect small phenotypic differences, even among conspecific populations [35].
Figure 2.
Overview of the morphometric measurements used in our study. Enumeration following Snoeks (2004). 1 = standard length (SL); 2 = body depth (BD); 3 = head length (HL); 4 = head width (HW); 5 = inter orbital width (IOW); 6 = snout length (SnL); 7 = lower jaw length (LJL); 8 = premaxillary pedicel length (PPL); 9 = cheek depth (ChD); 10 = eye diameter (ED); 11 = lacrimal depth (LaD); 16 = dorsal fin base length (DFB); 17 = anal fin base length (AFB); 18 = predorsal fin distance (PRD); 19 = preanal fin distance (PRA); 20 = prepectoral fin distance (PRP); 21 = preventral fin distance (PRV); 22 = caudal peduncle length (CPL); 23 = caudal peduncle depth (CPD).
Data were explored and analysed using principal component analysis on the log-transformed measurements. A covariance matrix was used to calculate the loadings of the variables and the scores of the specimens on the principal component axes. Loadings of the variables on the first principal component (PC1) were of the same sign and of a similar magnitude, indicating that this axis can be interpreted as a proxy for general size [36]. All computations were performed in the R software v. 2.11.1 [37].
3. Results
3.1. Genetic Differences
In C. sp. “Virginalis kajose,” allelic richness (AR) ranged from 16.46 (Chilumba) to 19.00 (Chenga Trawler), mean number of alleles per locus (Na) from 17.7 (Chilumba) to 21.7 (Mbenji Island), observed heterozygosity (HO) from 0.5929 (Nkhudzi Bay) to 0.7326 (Nkhotakota), and average expected heterozygosity (HE) from 0.7541 (Nkhata Bay) to 0.7854 (Senga Bay) for the Lake Malawi populations (Table 1). The population from Lake Malombe showed intermediate AR (16.92), Na (18.2), and HO (0.6195) and a lower HE (0.7375) compared to the Lake Malawi populations. In C. chrysonotus, all measures of genetic diversity (Na, AR, HE) except for HO were consistently lower in the Lake Malombe population compared to the Lake Malawi populations (Table 1).
Table 1.
Genetic diversity in two Copadichromis species, genotyped at six unlinked microsatellite loci. N: sample size; Na: mean number of alleles per locus (direct count); AR: allelic richness (i.e., standardized for minimal sample size 40 and 53); Ho: observed heterozygosity; He: unbiased expected heterozygosity. For each population, the P values for the single-locus tests for departure from Hardy-Weinberg equilibrium are tabulated. Populations: Chilumba (CH), Nkhata (NK), Nkhotakota (NO), Mbenji Island (MI), Senga Bay (SB), Chenga trawl (CT), Nkhudzi Bay (NKU), and Lake Malombe (LM).
(a) Copadichromis sp. “Virginalis kajose”
| Lake Malombe | Lake Malawi | |||||||
|---|---|---|---|---|---|---|---|---|
| CT | NKU | SB | MI | NO | NK | CH | ||
| N | 53 | 40 | 61 | 63 | 57 | 48 | 54 | 51 |
| Na | 18.2 | 19.0 | 19.8 | 20.5 | 21.7 | 20.0 | 18.7 | 17.7 |
| AR | 16.92 | 19.00 | 17.59 | 18.17 | 18.87 | 18.88 | 16.89 | 16.46 |
| Ho | 0.6195 | 0.7083 | 0.5929 | 0.7090 | 0.6871 | 0.7326 | 0.6821 | 0.683 |
| He | 0.7375 | 0.777 | 0.7729 | 0.7854 | 0.7648 | 0.7614 | 0.7541 | 0.7493 |
| Pzeb1 | <0.0001 | 0.0792 | <0.0001 | 0.0143 | 0.0179 | <0.0001 | 0.0021 | <0.0001 |
| Pzeb3 | 0.1075 | 0.2054 | <0.0001 | 0.0052 | 0.1802 | 0.8029 | 0.0306 | 0.3968 |
| Pzeb4 | 0.0160 | 0.4785 | 0.0141 | 0.0161 | <0.0001 | 0.2977 | 0.4464 | 0.0625 |
| Pzeb5 | 0.0001 | 0.0030 | <0.0001 | 0.6380 | 0.6631 | 0.1240 | 1.0000 | 0.3285 |
| UNH002 | <0.0001 | <0.0001 | 0.0068 | 0.4499 | <0.0001 | 1.0000 | 0.0295 | 0.0413 |
| TmoM11 | <0.0001 | 0.5857 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
(b) Copadichromis chrysonotus
| Lake Malombe | Lake Malawi | ||
|---|---|---|---|
| NO | CH | ||
| N | 54 | 65 | 53 |
| Na | 13.7 | 18.5 | 15.8 |
| AR | 13.61 | 17.51 | 15.80 |
| Ho | 0.6574 | 0.6718 | 0.6132 |
| He | 0.7265 | 0.7681 | 0.7776 |
| Pzeb1 | <0.0001 | 0.0029 | 0.0012 |
| Pzeb3 | 0.1486 | 0.1504 | <0.0001 |
| Pzeb4 | 0.6093 | 0.1347 | 0.0305 |
| Pzeb5 | 0.0104 | <0.0001 | <0.0001 |
| UNH002 | 0.0146 | 0.0503 | 0.0002 |
| TmoM11 | 0.5840 | 0.1782 | <0.0001 |
In C. sp. “Virginalis kajose,” 18 of 48 single-locus tests and in C. chrysonotus 7 of 18 single-locus tests for deviation from Hardy-Weinberg equilibrium were significant at a table-wide Bonferroni-corrected alpha level (αcorr = 0.001 and αcorr = 0.002 for the two species, resp.) (Table 1). Most deviations occurred across populations at loci Pzeb1, UNH002, and TmoM11, all involving heterozygote deficits. Loci Pzeb3, Pzeb4, and Pzeb5 were in agreement with Hardy-Weinberg equilibrium expectations in 6 of 33 single-locus tests (Table 1). Linkage disequilibrium tests across loci and populations revealed no significant allelic associations in C. sp. “Virginalis kajose” nor in C. chrysonotus (all P > .05).
Microsatellite data provided no evidence for a recent bottleneck in both species. We found no significant excess in gene diversity in any population. Concordantly, the mode-shift tests implemented in bottleneck showed no shift in the frequency distributions of microsatellite alleles. As expected for nonbottlenecked populations, all populations showed L-shaped allele frequencies.
In both species, we detected low but significant population structuring. The global measure for population differentiation across Lake Malombe populations was FST = 0.005 (P < .001) in C. sp. “Virginalis kajose” and FST = 0.008 (P < .001) in C. chrysonotus. Pairwise FST values are presented in Tables 2 and 3. For C. sp. “Virginalis kajose,” the measures for genetic differentiation between the Lake Malombe population and the populations from Lake Malawi were of the same order of magnitude as the genetic differentiation among populations within Lake Malawi (Table 2). For C. chrysonotus, the level of genetic differentiation between the Lake Malombe population and the Lake Malawi populations was higher than the level of genetic differentiation among the Lake Malawi populations (Table 3).
Table 2.
Pairwise values of FST (below the diagonal) and associated P values (above the diagonal) among eight C. sp. “Virginalis kajose” populations (codes following Table 1).
| Lake Malombe | Lake Malawi | |||||||
|---|---|---|---|---|---|---|---|---|
| CT | NKU | SB | MI | NO | NK | CH | ||
| LM | — | <0.0001 | 0.0021 | <0.0001 | <0.0001 | 0.2393 | 0.0002 | <0.0001 |
| CT | 0.009 | — | <0.0001 | 0.0253 | 0.0807 | 0.0002 | 0.0451 | <0.0001 |
| NKU | 0.005 | 0.007 | — | <0.0001 | <0.0001 | 0.0006 | 0.0178 | <0.0001 |
| SB | 0.007 | 0.001 | 0.007 | — | <0.0001 | 0.0055 | 0.0688 | <0.0001 |
| MI | 0.010 | 0.002 | 0.006 | 0.005 | — | <0.0001 | 0.0424 | <0.0001 |
| NO | 0.003 | 0.011 | 0.011 | 0.007 | 0.011 | — | <0.0001 | <0.0001 |
| NK | 0.006 | 0.007 | 0.004 | 0.001 | 0.002 | 0.008 | — | <0.0001 |
| CH | 0.011 | 0.010 | 0.015 | 0.010 | 0.008 | 0.013 | 0.009 | — |
Table 3.
Pairwise values of FST (below the diagonal) and associated P values (above the diagonal) among three C. chrysonotus populations (codes following Table 1).
| Lake Malombe | Lake Malawi | ||
|---|---|---|---|
| NO | CH | ||
| LM | — | <0.0001 | <0.0001 |
| NO | 0.01547 | — | <0.0001 |
| CH | 0.01430 | 0.00780 | — |
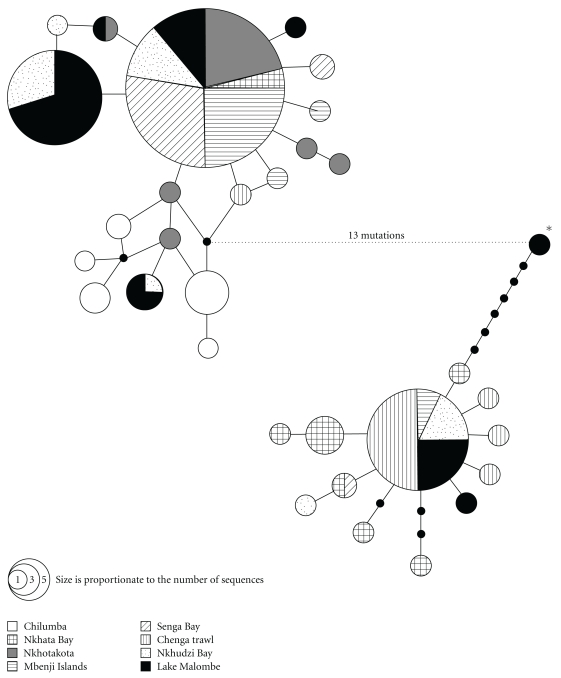
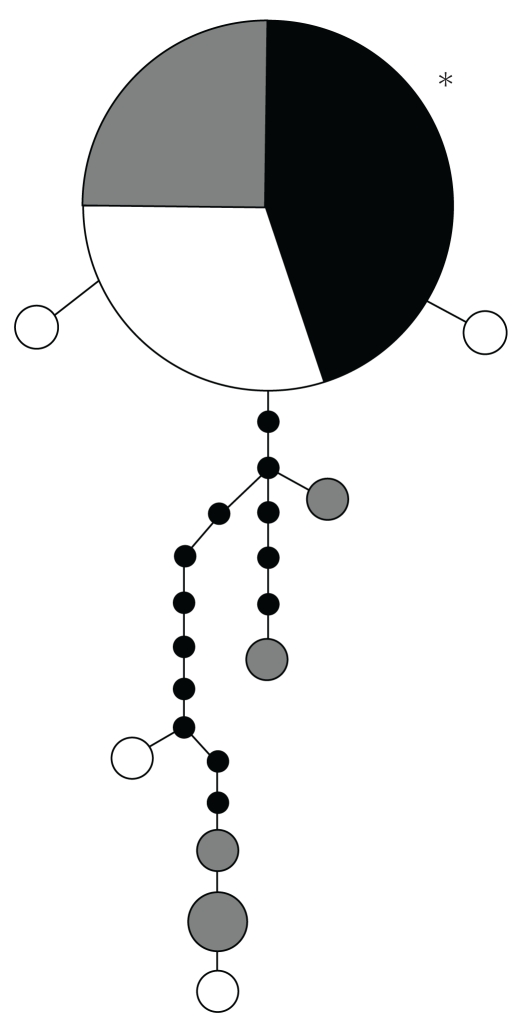
In the complete mtDNA dataset of 135 sequences, a total of 42 variable sites (ntotal = 325 bp) resulted in 40 haplotypes. The 98 sequences of C. sp. “Virginalis kajose” contained 36 polymorphic sites resulting in 32 haplotypes (Figure 3). The 37 C. chrysonotus sequences with 18 variable sites represented 9 haplotypes (Figure 4). Gene diversity in C. sp. “Virginalis kajose” ranged from 0.47 (Senga Bay) to 0.87 (Nkhudzi Bay) (Table 4). In C. chrysonotus, gene diversity was markedly lower ranging from zero (Lake Malombe) to 0.67 (Nkhotakota). Nucleotide diversity varied from 0.0037 (Nkhotakota) to 0.0263 (Nkhudzi Bay) in C. sp. “Virginalis kajose” and from zero (Lake Malombe) to 0.0180 (Nkhotakota) in C. chrysonotus. The only haplotype shared between the two species occurred in a single C. sp. “Virginalis kajose” from Lake Malombe (indicated with an asterisk in Figure 3) and in 28 C. chrysonotus from Lake Malombe (n = 13), Nkhotakota (n = 7,) and Chilumba (n = 8, indicated with an asterisk in Figure 4). Of the seven haplotypes found in C. sp. “Virginalis kajose” from Lake Malombe, six are also present in Lake Malawi, which contained 30 haplotypes in total. The haplotype that is unique to Lake Malombe differs by a single mutation (T to C) from a widespread haplotype, present in five populations from Lake Malawi.
Figure 3.
Haplotype network for the C. sp. “Virginalis kajose” sequences (based on 325 bp fragments of the mtDNA control region). The haplotype indicated with an asterisk is the haplotype that is shared with C. chrysonotus (see Figure 4).
Figure 4.
Haplotype network for the C. chrysonotus sequences (based on 325 bp fragments of the mtDNA control region). The haplotype indicated with an asterisk is the haplotype that is shared with C. sp. “Virginalis kajose” (see Figure 3). The legend is given in Figure 3.
Table 4.
Genetic diversity in two Copadichromis species based on a 325 bp mtDNA D-loop sequence. Sampling sites encoded as in Table 1. Genbank accession numbers for Copadichromis sp. “Virginalis kajose”: EF211832-211842, EF211848-211859, EF211868-211878, and EF647209-647272; Genbank accession numbers for C. chrysonotus: EF647273-647309, and EF647320-647331.
(a) Copadichromis sp. “Virginalis kajose”
| Lake Malombe | Lake Malawi | |||||||
|---|---|---|---|---|---|---|---|---|
| Population code | LM | CT | CH | NO | NK | MI | SB | NKU |
| Sample size | 21 | 10 | 12 | 11 | 10 | 12 | 11 | 11 |
| Number haplotypes | 7 | 5 | 5 | 6 | 7 | 6 | 3 | 6 |
| Number polymorphic sites | 22 | 17 | 6 | 5 | 22 | 15 | 15 | 19 |
| Gene diversity | 0.810 ± 0.004 | 0.667 ± 0.027 | 0.788 ± 0.008 | 0.727 ± 0.021 | 0.867 ± 0.011 | 0.682 ± 0.022 | 0.473 ± 0.026 | 0.873 ± 0.005 |
| Average pairwise difference | 6.981 | 3.4000 | 2.136 | 1.200 | 4.867 | 3.045 | 2.873 | 7.673 |
| Nucleotide diversity (average over loci) | 0.02161 ± 0.00002 | 0.01053 ± 0.00004 | 0.00659 ± 0.00001 | 0.00370 ± 0.00001 | 0.01507 ± 0.00005 | 0.00943 ± 0.00003 | 0.00889 ± 0.00004 | 0.02375 ± 0.00003 |
(b) Copadichromis chrysonotus
| Lake Malombe | Lake Malawi | ||
|---|---|---|---|
| Population code | LM | CH | NO |
| Sample size | 13 | 12 | 12 |
| Number haplotypes | 1 | 5 | 5 |
| Number polymorphic sites | 0 | 15 | 15 |
| Gene diversity | 0 | 0.576 ± 0.027 | 0.667 ± 0.020 |
| Average pairwise difference | 0 | 3.727 | 5.833 |
| Nucleotide diversity (average over loci) | 0 | 0.01150 ± 0.00003 | 0.0180 ± 0.00002 |
3.2. Body Size Differences
In both species, the specimens from Lake Malombe were smaller with an average SL of 55.1 ± 2.3 mm and 73.1 ± 6.8 mm for C. sp. “Virginalis kajose” and C. chrysonotus, respectively, compared to 90.2 ± 12.4 mm and 108.7 ± 11.4 mm in the populations from Lake Malawi.
3.3. Morphometric Differences
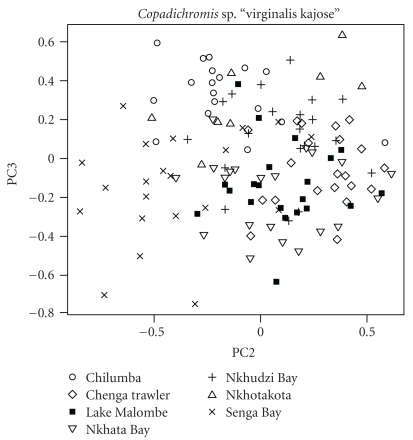
For C. sp. “Virginalis kajose,” PCA on the log-transformed measurements did not allow us to discriminate the Lake Malombe population from the Lake Malawi populations. The individual principal component scores overlapped largely on all PC axes for the specimens from both lakes (MANOVA, F(17,113) = 1.0889, P = .3731), except on PC1 which is a proxy for size. This overlap is illustrated for the scores on PC2 and PC3 in Figure 5. The variables that loaded most on PC2 were anal fin base length, caudal peduncle depth, body depth and eye diameter (Supplementary Table 1, see in Supplementary Material available at doi: 10.4061/2011/835946).
Figure 5.
Scatterplot of scores on the second and third axes of a PCA on log-transformed measurements for Copadichromis sp. “Virginalis kajose irginalis kajose”. The scores of the Lake Malombe individuals fall within the range of the scores for the Lake Malawi individuals.
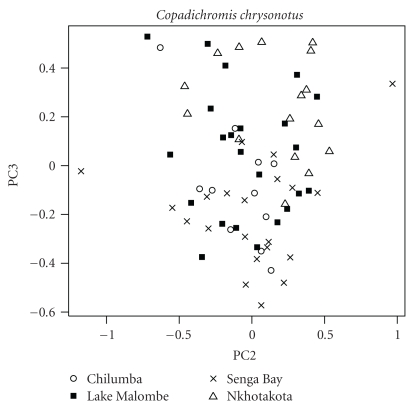
For C. chrysonotus, the specimens from Lake Malombe could not be distinguished from the Lake Malawi specimens by their scores on the principal component axes (MANOVA, F(17,53) = 0.4062, P = .9781; see Figure 6). The variables that loaded most with PC2 were eye diameter, anal fin base length, lower jaw length, body depth, and caudal peduncle depth (Supplementary Table 2).
Figure 6.
Scatterplot of scores on the second and third axes of a PCA on log-transformed measurements for Copadichromis chrysonotus. The scores of the Lake Malombe individuals fall within the range of the scores for the Lake Malawi individuals.
4. Discussion
4.1. Genetic Differentiation
Peripheral populations at the edge of a species' range are often less numerous than and isolated from core populations. Consequently, gene flow will be reduced, and stochastic demographic events such as bottlenecks, founder effects, and genetic drift may lead to a substantial loss of multilocus heterozygosity and allelic variation [1–3, 38]. In addition, elevated fishing mortality may also lead to a reduction in genetic variability perhaps owing to a selection response [39, 40], though at present the evidence of genetic change resulting from fishing is not unequivocal [41]. Populations of Copadichromis sp. Virginalis kajose and C. chrysonotus from Lake Malombe were expected to present reduced genetic variability when compared to their source populations in Lake Malawi because of their semi-isolated state, the distinct habitat conditions and elevated fishing pressure in the peripheral Lake Malombe.
We found low levels of population substructuring both within and among lakes, indicating that the studied fish species are relatively mobile and suggesting recent or ongoing gene flow between Lake Malawi and Lake Malombe populations. Lake Malombe is believed to have refilled around the 1930s [9], so the Lake Malombe population represents a relatively recent peripheral isolate. Furthermore, it has been shown that the Upper Shire River is used by various fish species migrating between the lakes [42], and this could probably also be the case for the two studied Copadichromis species. Despite the low degree of population subdivision, both species yielded a slightly decreased genetic diversity in Lake Malombe, especially for C. chrysonotus. We assume that the observed diminution in genetic variability is not caused by the high fishing mortality, because if true, we would rather expect a significant loss of genetic variation in C. sp. “Virginalis kajose” for which catch sizes in Lake Malombe have already declined [12]. Instead, a founder effect, that is, a population bottleneck associated with the colonization of Lake Malombe, may have been of principal importance for the observed lower genetic variation in these populations. Our microsatellite data did not however indicate for a heterozygosity excess nor a mode shift in the allele distribution pattern. Population bottlenecks of short duration are expected to substantially reduce the number of alleles present while having less effect on multilocus heterozygosity [43]. Moreover, our low sample sizes with respect to the high allelic richness may have lead to an artificial large proportion of low-frequency allele classes resulting in L-shaped allele frequency distributions and could therefore have influenced the robustness of the mode-shift test. Also, it is important to note that 18 of 48 single-locus tests in C. sp. “Virginalis kajose” and 7 of 18 in C. chrysonotus showed a deficit in heterozygotes. Most of the observed Hardy-Weinberg disequilibria were detected at three loci (Pzeb1, UNH002, and TmoM11) and are likely the result of the presence of nonamplifying null alleles (see [44] and references therein).
Inspection of the distribution pattern of mtDNA haplotypes provides support for the hypothesis of a natural founder effect with a different impact on each species. The single mtDNA haplotype detected in C. chrysonotus from Lake Malombe is the most common haplotype found for that species in Lake Malawi, with frequencies that exceed 58% in each of the sampled populations. Consequently, a stochastic shift in haplotype variation associated with the founding event, or genetic drift following colonization, may have resulted in the presence of only this haplotype in Lake Malombe for C. chrysonotus. Following the same reasoning, it appears logic that the change in haplotype variation was less pronounced for C. sp. “Virginalis kajose” because of the generally higher haplotype diversity in its source populations from Lake Malawi (Hmean = 0.725 versus Hmean = 0.520 for C. chrysonotus). Moreover, of the six haplotypes detected in the southernmost C. sp. “Virginalis kajose” population (Nkhudzi Bay), only the four most abundant ones are found in Lake Malombe.
4.2. Body Size Differences
Reduction in size at maturity is a predictive outcome of fishing pressure, either through fisheries-induced evolution or through phenotypic plasticity [45–47]. Reduction in body size was observed for heavily fished cichlid species, such as tilapia [48] and haplochromines from Lake Victoria [49, 50]. For both species in this study, the mean body size was significantly smaller in Lake Malombe than in Lake Malawi, which is in agreement with the hypothesis of a fishery pressure effect, although we cannot exclude the possibility that body size is affected by environmental effects. In any case, it is unlikely that the differences in body sizes are due to possible sampling artefacts, as our results are consistent with body sizes reported by Weyl et al. [12] from repetitive samples from Lake Malombe between March 2000 and April 2001.
4.3. Morphological Differentiation
Mayr [3] pointed out that populations at the edge of a species' range often diverge in morphology from central populations. Peripheral populations are likely to experience different regimes of ecological selection than central populations, often leading to adaptive divergence [4, 51]. In East African haplochromine cichlids, ecological adaptation is suggested to be one of the major mechanisms behind their explosive radiation [5, 52–54]. As such, smaller isolated peripheral water bodies may be of significant importance to the diversification of cichlid species in the East African Great Lakes by generating novel phenotypes in allopatry [3, 4, 8].
Lake Malombe's distinct habitat conditions (e.g., its shallow, eutrophic, turbid water state in contrast to the great visibility in the much deeper Lake Malawi [9]) would suggest an adequate basis for ecological differentiation leading to the generation of local phenotypes. Nonetheless, we found that peripheral populations of two Copadichromis species in Lake Malombe could not be distinguished morphologically from central populations in Lake Malawi. Theory predicts that adaptive divergence between populations reflects a balance between the diversifying effects of local selection and the homogenizing effects of gene flow [4, 55, 56]. Although morphological differentiation can occur in the face of gene flow, particularly when divergent selection is strong (see [57] and references therein), gene flow will often constrain morphological divergence [56]. The low degree of population subdivision as revealed by our microsatellite data seems to support the idea that gene flow between Lake Malombe and Lake Malawi populations is too substantial to allow for morphological divergence. Alternatively, it could also be that ecological divergence here is not manifested by divergence in shape, but rather by (e.g., physiological) adaptations which could not be perceived by our measurements.
5. Conclusion
Young peripheral populations of two Copadichromis species in Lake Malombe, connected to Lake Malawi via the Shire River, showed low genetic divergence and were morphometrically not distinct from their source populations in the main lake. This may be attributed to either homogenizing ongoing gene flow through the Shire River or very recently time since isolation of the populations in the satellite lake. Nonetheless, evidence was found for a founder event with a different impact on the genetic composition of the two species. We did not find direct evidence for a fisheries-induced impact on the genetic variability. Instead, body size for both species was significantly reduced in Lake Malombe, which could be a consequence of high fishing pressure and the use of small mesh sizes.
Supplementary Material
The supplementary material reports the loadings of the log-transformed metrics on the principal component axes for respectively C. sp. “Virginalis kajose” and C. chrysonotus. Loadings on PC1 are all very similar, indicating that PC1 is a proxy for size. Most variation in shape is represented by PC2. For C. “Virginalis kajose”, the anal fin base length, caudal peduncle length, body depth and eye diameter are the metrics that showed the highest loadings on PC2. The metrics that loaded most with PC2 for C. chrysonotus were eye diameter, anal fin base length, lower jaw length, body depth and caudal peduncle depth.
Acknowledgments
The field work was supported by the Interdisciplinary Research Centre of the K.U.Leuven Campus Kortrijk, a grant from the King Leopold III Fund for Nature Exploration and Conservation to D. Anseeuw, grants from the Research Foundation Flanders (FWO), and the “Stichting tot bevordering van het wetenschappelijk onderzoek in Afrika” to J. Snoeks. We wish to thank the Malawi Fisheries Department, and especially the late Sam Mapila and Orton Kachinjika, for permission to conduct fieldwork in Malawi, the Department for International Development (DFID) of the British High Commission in Malawi and especially George Turner (University of Hull; presently at Bangor University), for putting a 4 × 4 vehicle at our disposal for the sampling trip. The authors thank the late Davis Mandere for his valuable assistance in the field, the Fisheries Research Unit in Monkey Bay and Nkhata Bay for their assistance and the late Stuart Grant for his hospitality. The authors thank professor Lea Vermeire for her statistical support.
References
- 1.Lesica P, Allendorf FW. When are peripheral populations valuable for conservation? Conservation Biology. 1995;9(4):753–760. [Google Scholar]
- 2.Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29(1):1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 3.Mayr E. Change of genetic environment and evolution. In: Huxley J, Hardy AC, Ford EB, editors. Evolution as a Process. London, UK: Allen & Unwin; 1954. [Google Scholar]
- 4.García-Ramos G, Kirkpatrick M. Genetic models of adaptation and gene flow in peripheral populations. Evolution. 1997;51(1):21–28. doi: 10.1111/j.1558-5646.1997.tb02384.x. [DOI] [PubMed] [Google Scholar]
- 5.Salzburger W, Meyer A. The species flocks of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften. 2004;91(6):277–290. doi: 10.1007/s00114-004-0528-6. [DOI] [PubMed] [Google Scholar]
- 6.Seehausen O. African cichlid fish: a model system in adaptive radiation research. Proceedings of the Royal Society B. 2006;273(1597):1987–1998. doi: 10.1098/rspb.2006.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenwood PH. The cichlid fishes of Lake Nabugabo, Uganda. Bulletin of the British Museum (Natural History. Zoology. 1965;12:315–357. [Google Scholar]
- 8.Genner MJ, Nichols P, Carvalho GR, et al. Evolution of a cichlid fish in a Lake Malawi satellite lake. Proceedings of the Royal Society B. 2007;274(1623):2249–2257. doi: 10.1098/rspb.2007.0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tweddle D, Turner GF, Seisay BD. Changes in species composition and abundance as a consequence if fishing in Lake Malombe, Malawi. In: Pitcher TJ, Hart JB, editors. The Impact of Species Changes in African Great Lakes. London, UK: Chapman and Hall; 1995. pp. 413–424. [Google Scholar]
- 10.Weyl OLF. Small-scale fisheries statistics summary. National Aquatic Resource Management Programme (NARMAP), Short communication, 2001.
- 11.Weyl OLF, Banda MC, Manase W, Namoto W, Mwenekibombwe LH. Analysis of catch and effort data for the fisheries of Lake Malombe, 1976–1999. Government of Malawi. Fisheries Bulletin. 2001;45:p. 34. [Google Scholar]
- 12.Weyl OLF, Mwakiyongo KR, Mandere DS. An assessment of the nkacha net fishery of Lake Malombe, Malawi. African Journal of Aquatic Science. 2004;29(1):47–55. [Google Scholar]
- 13.Emerson BC. Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Molecular Ecology. 2002;11(6):951–966. doi: 10.1046/j.1365-294x.2002.01507.x. [DOI] [PubMed] [Google Scholar]
- 14.Hauser L, Adcock GJ, Smith PJ, Ramírez JHB, Carvalho GR. Loss of microsatellite diversity and low effective population size in an overexploited population of New Zealand snapper (Pagrus auratus) Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11742–11747. doi: 10.1073/pnas.172242899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birky CW, Maruyama T, Fuerst P. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics. 1983;103(3):513–527. doi: 10.1093/genetics/103.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarne P, Lagoda PJL. Microsatellites, from molecules to populations and back. Trends in Ecology and Evolution. 1996;11(10):424–429. doi: 10.1016/0169-5347(96)10049-5. [DOI] [PubMed] [Google Scholar]
- 17.Witte F, Welten M, Heemskerk M, et al. Major morphological changes in a Lake Victoria cichlid fish within two decades. Biological Journal of the Linnean Society. 2008;94(1):41–52. [Google Scholar]
- 18.Kishe-Machumu M, Witte F, Wanink JH. Dietary shift in benthivorous cichlids after the ecological changes in Lake Victoria. Animal Biology. 2008;58(4):401–417. [Google Scholar]
- 19.Rutjes HA, De Zeeuw MP, van den Thillart GEEJM, Witte F. Changes in ventral head width, a discriminating shape factor among African cichlids, can be induced by chronic hypoxia. Biological Journal of the Linnean Society. 2009;98(3):608–619. [Google Scholar]
- 20.Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research. 1997;25(22):4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Oppen MJH, Rico C, Deutsch JC, Turner GF, Hewitt GM. Isolation and characterization of microsatellite loci in the cichlid fish Pseudotropheus zebra. Molecular Ecology. 1997;6(4):387–388. doi: 10.1046/j.1365-294x.1997.00188.x. [DOI] [PubMed] [Google Scholar]
- 22.Kellogg KA, Markert JA, Stauffer JR, Kocher TD. Microsatellite variation demonstrates multiple paternity in lekking cichlid fishes from Lake Malawi, Africa. Proceedings of the Royal Society B. 1995;260(1357):79–84. [Google Scholar]
- 23.Zardoya R, Vollmer DM, Craddock C, Streelman JT, Karl S, Meyer A. Evolutionary conservation of microsatellite flanking regions and their use in resolving the phylogeny of cichlid fishes (Pisces: Perciformes) Proceedings of the Royal Society B. 1996;263(1376):1589–1598. doi: 10.1098/rspb.1996.0233. [DOI] [PubMed] [Google Scholar]
- 24.Meyer A, Kocher TD, Basasibwaki P, Wilson AC. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature. 1990;347(6293):550–553. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- 25.Belkhir K. GENETIX, Version 4.05. Montpellier, France: Laboratoire Genome, Populations, Interactions, CNRS UPR 9060; 2000. [Google Scholar]
- 26.Raymond M, Rousset F. GENEPOP version 1.2: populations genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- 27.Wright S. Evolution and the Genetics of Populations. Vol. 2: The Theory of Gene Frequencies. Chicago, Ill, USA: University of Chicago Press; 1969. [Google Scholar]
- 28.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 29.Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001, http://www2.unil.ch/popgen/softwares/fstat.htm.
- 30.Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144(4):2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luikart G, Cornuet JM. Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology. 1998;12(1):228–237. [Google Scholar]
- 32.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, Version 4.10.8. Barcelona, Spain: Universitat de Barcelona; 2006. [Google Scholar]
- 34.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 35.Snoeks J. The Cichlid Diversity of Lake Malawi/Nyassa/Niassa: Identification, Distribution and Taxonomy. Big Bend, Tex, USA: Cichlid Press; 2004. [Google Scholar]
- 36.Bookstein F. Morphometric Tools for Landmark Data. Geometry and Biology. Cambridge, UK: Cambridge University Press; 1991. [Google Scholar]
- 37.R Development Core Team. R: a language and environment for statistical computing. R foundation for statistical computing. Vienna, Austria, 2010, http://www.r-project.org/
- 38.Lande R. Genetic variation and phenotypic evolution during allopatric speciation. The American Naturalist. 1980;116:463–479. [Google Scholar]
- 39.Quinn TP, Hodgson S, Flynn L, Hilborn R, Rogers DE. Directional selection by fisheries and the timing of sockeye salmon (Oncorhynchus nerka) migrations. Ecological Applications. 2007;17(3):731–739. doi: 10.1890/06-0771. [DOI] [PubMed] [Google Scholar]
- 40.Swain DP, Sinclair AF, Mark Hanson J. Evolutionary response to size-selective mortality in an exploited fish population. Proceedings of the Royal Society B. 2007;274(1613):1015–1022. doi: 10.1098/rspb.2006.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hutchings JA, Fraser DJ. The nature of fisheries- and farming-induced evolution. Molecular Ecology. 2008;17(1):294–313. doi: 10.1111/j.1365-294X.2007.03485.x. [DOI] [PubMed] [Google Scholar]
- 42.FAO. CIFA Technical Paper. 21. Rome, Italy: FAO; 1993. Fisheries Management in the south-east arm of Lake Malawi, the Upper Shire River and Lake Malombe, with particular reference to the fisheries on chambo (Oreochromis spp.) [Google Scholar]
- 43.Allendorf FW. Genetic drift and the loss of alleles versus heterozygosity. Zoological Biology. 1986;5:181–190. [Google Scholar]
- 44.Anseeuw D, Maes GE, Busselen P, Knapen D, Snoeks J, Verheyen E. Subtle population structure and male-biased dispersal in two Copadichromis species (Teleostei, Cichlidae) from Lake Malawi, East Africa. Hydrobiologia. 2008;615(1):69–79. [Google Scholar]
- 45.Law R. Fishing, selection, and phenotypic evolution. ICES Journal of Marine Science. 2000;57(3):659–668. [Google Scholar]
- 46.Reznick DN, Ghalambor CK. Can commercial fishing cause evolution? Answers from guppies (Poecilia reticulata) Canadian Journal of Fisheries and Aquatic Sciences. 2005;62(4):791–801. [Google Scholar]
- 47.Kuparinen A, Merilä J. Detecting and managing fisheries-induced evolution. Trends in Ecology and Evolution. 2007;22(12):652–659. doi: 10.1016/j.tree.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 48.Blay J, Asabereameyaw A. Assessment of the fishery of a stunted population of the cichlid, Sarotherodon melanotheron (Ruppel), in a closed lagoon in Ghana. Journal of Applied Ichthyology. 1993;9:1–11. [Google Scholar]
- 49.Witte F, Barel KDN, van Oijen MJP. Intraspecific variation of haplochromine cichlids from Lake Victoria and its taxonomic implications. South African Journal of Science. 1997;93(11-12):585–594. [Google Scholar]
- 50.Schwartz JDM, Pallin MJ, Michener RH, Mbabazi D, Kaufman L. Effects of Nile perch, Lates niloticus, on functional and specific fish diversity in Uganda’s Lake Kyoga system. African Journal of Ecology. 2006;44(2):145–156. [Google Scholar]
- 51.Endler JA. Geographic Variation, Speciation and Clines. Princeton, NJ, USA: Princeton University Press; 1977. [PubMed] [Google Scholar]
- 52.Liem KF. Evolutionary strategies and morphological innovations—cichlid pharyngeal jaws. Systematic Zoology. 1973;189:93–125. [Google Scholar]
- 53.Danley PD, Kocher TD. Speciation in rapidly diverging systems: lessons from Lake Malawi. Molecular Ecology. 2001;10(5):1075–1086. doi: 10.1046/j.1365-294x.2001.01283.x. [DOI] [PubMed] [Google Scholar]
- 54.Hulsey CD. Cichlid genomics and phenotypic diversity in a comparative context. Integrative and Comparative Biology. 2009;49(6):618–629. doi: 10.1093/icb/icp071. [DOI] [PubMed] [Google Scholar]
- 55.Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236(4803):787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- 56.Hendry AP, Taylor EB. How much of the variation in adaptive divergence can be explained by gene flow? An evaluation using lake-stream stickleback pairs. Evolution. 2004;58(10):2319–2331. doi: 10.1111/j.0014-3820.2004.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 57.Garant D, Forde SE, Hendry AP. The multifarious effects of dispersal and gene flow on contemporary adaptation. Functional Ecology. 2007;21(3):434–443. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary material reports the loadings of the log-transformed metrics on the principal component axes for respectively C. sp. “Virginalis kajose” and C. chrysonotus. Loadings on PC1 are all very similar, indicating that PC1 is a proxy for size. Most variation in shape is represented by PC2. For C. “Virginalis kajose”, the anal fin base length, caudal peduncle length, body depth and eye diameter are the metrics that showed the highest loadings on PC2. The metrics that loaded most with PC2 for C. chrysonotus were eye diameter, anal fin base length, lower jaw length, body depth and caudal peduncle depth.