Abstract
Background and Aims
Polyploidy is a dominant feature of flowering-plant genomes, including those of many important crop species. Arachis is a largely diploid genus with just four polyploid species. Two of them are economically important: the cultivated peanut and A. glabrata, a tropical forage crop. Even though it is usually accepted that polyploids within papilionoid legumes have arisen via hybridization and further chromosome doubling, it has been recently suggested that peanut arose through bilateral sexual polyploidization. In this paper, the polyploid nature of the recent, spontaneously originated triploid cytotype of the tropical lucerne, A. pintoi, was analysed, and thereby the mechanism by which polyploids may arise in the genus.
Methods
Chromosome morphology of 2x and 3x A. pintoi was determined by the Feulgeńs technique and the rDNA sites were mapped by FISH. To investigate whether polyploidization occurred by means of unreduced gametes, a detailed analysis of the microsporogenesis and pollen grains was made.
Key Results
The 2x and 3x plants presented 9m + 1sm and a satellited chromosome type 2 in each haploid genome. Physical mapping revealed a cluster of 18S–26S rDNA, proximally located on chromosome 6, and two 5S rDNA loci on chromosomes 3 and 5. Diploid plants presented 10II in meiosis while trivalents were observed in all triploids, with a maximum of 10III by cell. Diploid A. pintoi produced normal tetrads, but also triads, dyads and monads. Two types of pollen grains were detected: (1) normal-sized with a prolate shape and (2) large ones with a tetrahedral morphology.
Conclusions
Karyotype and meiotic analysis demonstrate that the 3x clone of A. pintoi arose by autopolyploidy. The occurrence of unreduced gametes strongly supports unilateral sexual polyploidization as the most probable mechanism that could have led to the origin of the triploid cytotype. This mechanism of polyploidization would probably be one of the most important mechanisms involved in the origin of economically important species of Arachis, either by triploid bridge or bilateral sexual polyploidization.
Keywords: Arachis pintoi, autotriploid, Caulorrhizae, chromosome pairing, fluorescent in situ hybridization, unreduced gametes
INTRODUCTION
The genus Arachis (Leguminosae) is native to South America and has 80 formally recognized species (Krapovickas and Gregory, 1994; Valls and Simpson, 2005). These entities have been assembled into nine sections according to morphology, geographic distribution and cross compatibility. Among them, the Caulorrhizae section includes two species: A. pintoi and A. repens. These species are diploids with 2n = 2x = 20 (Fernández and Krapovickas, 1994) and grow naturally in the states of Goiás, Bahia and Minas Gerais in Brazil. Arachis pintoi, besides having good seed set in most accessions, is adapted for clonal propagation by means of stolons. Arachis repens rarely sets seeds but is easily propagated from stolons.
Arachis pintoi produces high quality forage (Grof, 1985), and several cultivars have been released in Colombia and Brazil. Among them ‘Maní Forrajero Perenne’ (Rincón et al., 1992) is the most widely cultivated in the tropics and is usually referred to as the tropical lucerne. Many cultivars and wild accession of this species are highly adapted to acid and low fertility soils, have a high nutritive value, and persist among aggressive grasses like Brachiaria and Panicum (Lascano and Thomas, 1988)
A clone with higher forage productivity was isolated from a cultivated plot of diploid A. pintoi ‘Maní Forrajero Perenne’ in Costa Rica. Chromosome counts on material from this clone have shown that it is triploid (Peñaloza et al., 1996), but the clone has not been analysed further. In spite of this paucity of data, the triploid cytotype is largely accepted as having arisen by autopolyploidy, but there is no evidence in this regard. Therefore, the first objective of this study was to determine the nature of this triploid accession of A. pintoi.
One of the ways to determine the genome constitution of polyploids is by analysing karyotypes using classical techniques, and for those groups with small, similar chromosomes, homologies have been established by physical mapping of rDNA genes (Jiang and Gill, 1994). Particularly, in Arachis species, the ability to detect ribosomal genes by FISH has increased the number of chromosomal markers, allowing the establishment of homologies for more chromosomes than was possible with classical cytogenetics (Robledo and Seijo, 2008; Robledo et al., 2009).
Another powerful tool to investigate whether a species is an auto- or allopolyploid is by analysing meiotic behaviour and determining meiotic chromosomal associations during diakinesis or metaphase I (Jauhar and Joppa, 1996). According to the number and type of chromosome associations (univalents, bivalents, trivalents, tetravalents), it is possible to determine the polyploid status of the species under study (Stebbins, 1947).
Therefore, to attain the first objective, the following were analysed: (a) the chromosome homologies between the 2x and 3x cytotypes of A. pintoi by determining the karyotype formulae using Feulgen's technique and by mapping the 5S and the 18S–25S rDNA sites using FISH; and (b) the degree of homology of the triploid chromosome complements by establishing the meiotic configurations during meiotic metaphase I.
In the genus Arachis, there are four polyploid species. Two are economically important: cultivated peanut, Arachis hypogaea, an allotetraploid that originated around 5000 years bp (Bonavia, 1982); and A. glabrata, a forage crop in the southern United States and Australia. Even though A. glabrata has been considered to be an autopolypoid with different degrees of diploidization, its polyploid nature is still unknown. Arachis hypogaea, is usually accepted to have arisen via hybridization and further chromosome doubling. However, recently we proposed that this cultigen arose through bilateral sexual polyploidization as the mechanism of origin for this cultigen (Seijo et al., 2007). The recent, spontaneous origin of the triploid A. pintoi provides a unique opportunity to study the mechanism by which polyploids may arise in the genus and the genomic processes that occurred after that event.
The analysis of meiotic behaviour, sporads and pollen grains in other plant groups has revealed the occurrence of 2n pollen and the possibility that new cytotypes arise by sexual polyploidization (Bretagnolle, 2001; Jauhar, 2007). In the genus Arachis, the formation of unreduced gametes has been reported in a triploid artificial hybrid, A. hypogaea × (A. cardenasii × A. diogoi) (Simpson and Davis, 1983); however, there are no records of such an occurrence in diploid taxa.
In this context, the second objective was to obtain evidence to understand the mechanism involved in the origin of the triploid A. pintoi to infer the mechanism of polyploid formation in Arachis as a whole. For this purpose, the microsporogenesis and pollen grains (morphology and size) of the diploid accession were analysed in the search for 2n gametes.
MATERIALS AND METHODS
Plant material
The material studied was the cultivar Arachis pintoi Krapov. & W.C. Gregory ‘Maní Forrajero Perenne’: CIAT 17434 with diploid cytotype (2n = 2x = 20), derived from the original botanical collection, Gregory and Krapovickas 12787; and triploid plants (2n = 3x = 30), originally from a plot of diploid ‘Maní Forrajero Perenne’ from Costa Rica. Both materials are in the germplasm bank of the Instituto de Botánica del Nordeste, Corrientes, Argentina.
Chromosome preparations and Feulgen staining in 2x and 3x A. pintoi
Plants of the two cytotypes were obtained from stolons and grown in pots. Roots 10–15 mm long were directly collected from those plants, pretreated with 2 mm L−1 8-hydroxyquinoline for 3 h at room temperature (Fernández and Krapovickas, 1994) and then fixed in 3 : 1 absolute ethanol : glacial acetic acid for a minimum of 12 h at 4 °C. Chromosomes were stained using Feulgen's technique, then squashed in a drop of 2 % acetic orcein.
Fluorescent in situ hybridization in 2x and 3x A. pintoi
Chromosome spreads
Until fixation, the steps were the same as those just described, then somatic chromosome spreads were prepared according to Schwarzacher et al. (1980). Root apices were digested in 1 % (w/v) cellulose (from Trichoderma viridae, Onozuka R-10; Serva, Heidelberg, Germany) plus 10 % (v/v) pectinase dissolved in 40 % glycerol (from Aspergillus niger; Sigma, St Louis, Missouri, USA) in 0·01 m L−1 citrate buffer, pH 4·8, at 37 °C for 2 h. Subsequently, the meristematic cells were removed from the root tips and squashed in 45 % acetic acid. After removal of the coverslip with carbon dioxide, the slides were air-dried, aged for 1–2 d at room temperature, and then kept at −20 °C until use.
Probe labelling and fluorescent in situ hybridization
The 5S rDNA and 18S–26S rDNA loci were localized using probes pA5S, pA18S and pA26S isolated from genomic DNA of A. hypogaea (Robledo and Seijo, 2008) and labelled by nick translation with digoxigenin-11-dUTP (Boehringer Mannheim, Mannheim, Germany) or biotin-11-dUTP (Sigma). Pretreatment of slides, chromosome and probe denaturation, conditions for the in situ hybridization (hybridization mixes contained DNA probes at a concentration of 2·5–3·5 ng μL−1, with a stringency to allow sequences with 80–85 % identity to remain hybridized), post-hybridization washing, blocking and indirect detection with fluorochrome-conjugated antibodies were performed according to Moscone et al. (1996). The first set of antibodies consisted of mouse anti-biotin (Dakopatts, Dako, Carpinteria, CA, USA) and sheep anti-digoxigenin conjugated to fluorescein isothiocyanate (FITC) (Boehringer Mannheim). The second set of antibodies consisted of rabbit anti-mouse conjugated to tetramethyl-rhodamine isothiocyanate (TRITC) (Dakopatts) and FITC-conjugated rabbit antisheep (Dakopatts). Preparations were counterstained and mounted with Vectashield medium (Vector Laboratories, Burlingame, CA, USA) containing 2 mg mL−1 of 4′,6-diamidino-2-phenylindole (DAPI).
Fluorescence microscopy and image acquisition
Chromosomes were viewed and photographed with a Leica DMRX fluorescence microscope (Leica, Heerbrugg, Switzerland) equipped with a computer-assisted Leica DC 350 digital camera system. Red, green, and blue images were captured in black and white using appropriate filters for TRITC, FITC and DAPI excitation, respectively. Digital images were processed with Photoshop, version 7·0 (Adobe, San Jose, CA, USA).
Meiotic analysis
Inflorescences of 2x and 3x A. pintoi were fixed in 3 : 1 absolute ethanol:acid acetic for 24 h, transferred to 70 % ethanol and refrigerated until use. Pollen mother cells (PMCs) were then stained in a drop of 2 % lactopropionic orcein. Representative permanent slides were made using Euparal as mounting medium.
Production of 2n gametes in 2x A. pintoi
Sporads were screened, and the occurrence of 2n gametes in dyads and triads was recorded. Unreduced pollen grains were identified by a detailed examination of pollen throughout the range of sizes produced by each individual (Bretagnolle and Thompson, 1995). This method is based on the fact that cell (pollen) size increases as DNA content increases (Bennett, 1972). Pollen samples were obtained from two flowers (each containing a mixture of pollen grains from different anthers) in each of the five individual plants analysed and stained by 1 : 2 % acetocarmine : glycerin. These slides were used for estimating the pollen viability and to determine the mean pollen size.
Analysis of pollen-grain morphology by scanning electron microscopy (SEM)
Pollen samples were obtained from fresh flowers of plants grown under greenhouse conditions. To analyse pollen shape, grains were acetolysed according to the procedure suggested by Erdtman (1966). The acetolysed grains were first washed in 96 % alcohol, and then in absolute alcohol. The samples were air-dried in Petri dishes, sputtered with gold–palladium and finally observed with a JEOL 5800 LV scanning electron microscope at 15 kV. The terminology used to describe pollen follows Punt et al. (2007).
RESULTS
Karyotypic features of 2x and 3x A. pintoi
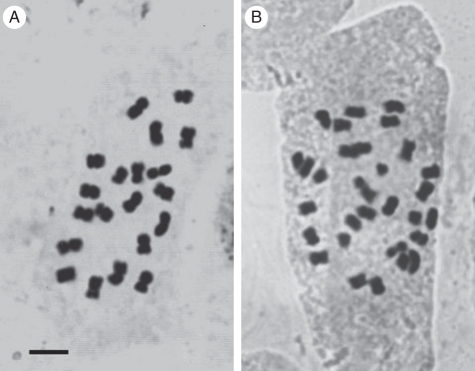
Classical cytological analysis demonstrated that the diploid clone has 18 metacentrics (m) and two submetacentrics (sm), while the triploid presents 30 chromosomes (Fig. 1) arranged in 27m + 3sm. Two satellited chromosomes (SAT chromosomes) type 2 were observed in the diploid and three in the triploid.
Fig. 1.
Somatic chromosomes of A. pintoi with Feulgen's staining: (A) diploid cytotype with 2n = 20 chromosomes; (B) triploid cytotype with 2n = 30 chromosomes. Scale bar = 5 µm.
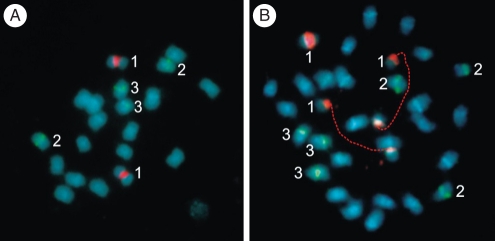
Physical mapping of rDNA genes using FISH revealed clusters of 18S–26S rDNA exclusively localized in the secondary constriction of the SAT chromosomes; these signals were located near the centromere on the short arms of metacentric pair 6 in both the 2x and the 3x plants (Fig. 2). In all metaphases, 18S–26S rDNA loci presented fluorescent signals of similar intensity. In situ hybridization with the 5S rDNA probe revealed two signals of similar intensity per haploid complement. These loci were localized on the short arms of metacentric chromosomes 3 and 5, both in diploid (Fig. 2A) and triploid plants (Fig. 2B).
Fig. 2.
Somatic chromosomes of (A) 2x and (B) 3x plants of A. pintoi after double fluorescent in situ hybridization (FISH) showing yellow-green fluorescein isothiocyanate (FITC) signals from 5S rDNA probe (two chromosomes per haploid complement, indicated by numbers 2 and 3) and red tetramethyl-rhodamine isothiocyanate (TRITC) signals from the 45S rDNA probe (indicated by number 1). Chromosomes were counterstained with DAPI (light blue) after the FISH. Homologous chromosomes were indicated with same numbers.
The 2x clones always had two chromosomes with active 18S–26S rDNA clusters (i.e. those that in Arachis characteristically remained uncondensed at metaphase), while the number in the 3x plants varied between individuals or even between cells of the same plant. Three active clusters were observed in 31 % of the cells, two in 61 % and one in 8 % of them.
Microsporogenesis in triploid plants
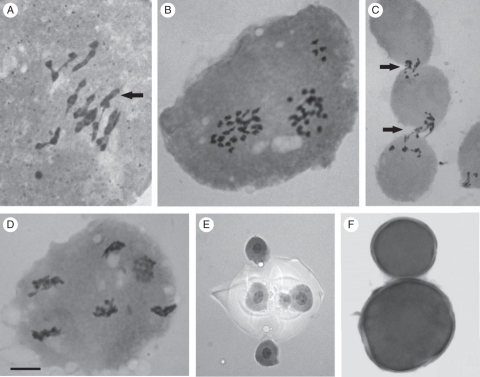
Different configurations in the PMCs at diakinesis/metaphase I were found (Table 1); 8III + 2II + 2I was the most frequent (in approx. 40 % of the cells). Trivalents were observed in 100 % of the analysed cells in diakinesis/metaphase I at frequencies that ranged from one to ten per cell (Fig. 3A). The percentage of cells with ten trivalents was 18·75 % (Table 1), and the mean frequency of trivalents per cell was 8·67 (Table 2).
Table 1.
Meiotic behaviour of 3x A. pintoi in different meiotic phases (metaphase I, anaphase I, telophase I, prophase II, metaphase II and anaphase II) and sporad analysis
| Phase | No. of PMCs | % |
|---|---|---|
| Metaphase I | ||
| 10III | 3 | 18·75 |
| 9III + 1II + 1I | 5 | 31·25 |
| 8III + 2II + 2I | 7 | 43·75 |
| Out-of-plate chromosomes | 1 | 6·25 |
| Anaphase I | ||
| Normal (regular segregation of chromosomes) | 3 | 33·33 |
| With one laggard chromosome | 3 | 33·33 |
| With two laggard chromosomes | 2 | 22·22 |
| With five laggard chromosomes | 1 | 11·11 |
| Telophase I | ||
| Normal (all chromosomes in two nuclei) | 15 | 99·99 |
| Out-of-nucleus chromosomes | 1 | 0·01 |
| Prophase II (number of chromosomes per nucleus) | ||
| 15/15 | 17 | 58·62 |
| 14/16 | 8 | 27·58 |
| 13/17 | 1 | 3·45 |
| 18/12 | 1 | 3·45 |
| 19/11 | 2 | 6·90 |
| Metaphase II | ||
| Normal (all chromosomes in two plates) | 7 | 77·77 |
| Out-of- plate chromosome (1) | 1 | 11·11 |
| Out-of- plate chromosomes (2) | 1 | 11·11 |
| Anaphase II | ||
| Normal (all chromosomes segregate in two spindles) | 18 | 81·82 |
| With four laggard chromosomes | 1 | 4·54 |
| With two laggard chromosomes | 1 | 4·54 |
| With six poles | 2 | 9·10 |
| Sporads | ||
| Tetrads | 362 | 97·05 |
| Tetrads with two larger size microspores | 1 | 0·27 |
| Tetrads with micromicrospore | 8 | 2·14 |
| Triads | 1 | 0·27 |
| Dyads | 1 | 0·27 |
Fig. 3.
Microsporogenesis and pollen grains in 3x plants of A. pintoi: (A) meiotic behaviour in metaphase I, 9III + 1II + 1I (arrow indicates the bivalent); (B) four chromosomes out of the prophase II nuclei; (C) cytomixis (arrows indicate chromatin migration between PMCs through cytoplasmatic channels); (D) six poles in telophase II; (E) pentad with a micromicrospore; (F) two different sizes of pollen grains (large and normal pollen). The scale bar in (D) applies to all the images, as follows: (A, B, D) = 5 µm; (C, E) = 7·5 µm; (F) = 15 µm.
Table 2.
Meiotic chromosome associations, mean, range of variation and standard error (s.e.) in triploid cytotype of A. pintoi
| Statistic | Univalents | Bilavents | Trivalents |
|---|---|---|---|
| Mean | 1·33 | 1·33 | 8·67 |
| Range | 0–2 | 0–2 | 8–10 |
| s.e. | 0·22 | 0·22 | 0·23 |
Meiotic irregularities such as out-of-plate and out-of-nuclei chromosomes, laggard chromosomes, and cytomixis (i.e. migration of chromatin between meiocytes through cytoplasmic channels) were observed in different phases of meiosis I and II (Fig. 3B–D) and are summarized in Table 1. Segregation at anaphase I and II was irregular, leading to the formation of unbalanced gametes (Fig. 3D–E). Variation in pollen size and a high percentage of unstained pollen (57·53 %) was recorded in the triploid clone after carmine–glycerine staining (Fig. 3F).
Microsporogenesis of the diploid plants
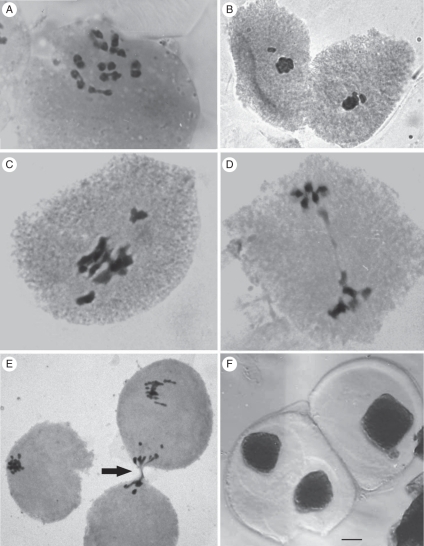
Meiotic analysis of the diploid cytotype showed the presence of 10II per cell (Fig. 4A) in all the PMCs at metaphase I. Although bivalent formation was regular, some irregular phenomena were observed in cells of different phases (Table 3). Out-of-plate bivalents were found in 14·49 % of the metaphase I cells (Fig. 4B), and chromosome bridges were observed in 0·99 % of cells in ana-telophase I (Fig. 4D). Cytomictic processes were observed in 5·80 % of the metaphases and in 2·97 % of the telophases I (Fig. 4E). The sporads analysis showed that, in addition to the expected normal tetrads, triads, dyads and monads were formed as a result of the meiotic division (Fig. 4F).
Fig. 4.
Microsporogenesis in diploid plants of A. pintoi: (A) metaphase I with 10II; (B) out-of-plate chromosomes in metaphase I; (C) metaphase I showing bivalents with early migration to the poles; (D) chromosome bridge in telophase I; (E) cytomixis (arrow indicates chromatin migration between PMCs through cytoplasmatic channels); (F) dyad (left) and monad (right). The scale bar in (F) applies to all the images, as follows: (A, B, D) = 5 µm; (C, E) = 7·5 µm; (F) = 15 µm.
Table 3.
Meiotic behaviour of 2x A. pintoi in different phases (metaphase I, telophase I, telophase II) and sporad analysis
| Phase | No. of CMPs | % |
|---|---|---|
| Metaphase I | ||
| 20II | 55 | 79·71 |
| Out-of-plate bivalent | 10 | 14·49 |
| Cytomixis | 4 | 5·80 |
| Telophase I | ||
| Normal (all chromosomes in two nuclei) | 97 | 96·04 |
| Bridge in anaphase | 1 | 0·99 |
| Cytomixis | 3 | 2·97 |
| Telophase II | ||
| Normal (all chromosomes in four nuclei) | 20 | 100 |
| Sporads | ||
| Tetrad | 399 | 91·94 |
| Triads | 2 | 0·46 |
| Dyads | 2 | 0·46 |
| Monads | 31 | 7·14 |
Pollen grain analysis by SEM revealed that pollen grains were a mixture of 91·34 % prolate tricolpate reticulate grains (Fig. 5A), 6·87 % of tetrahedral grains (Fig. 5B) and 1·79 % of unviable grains.
Fig. 5.
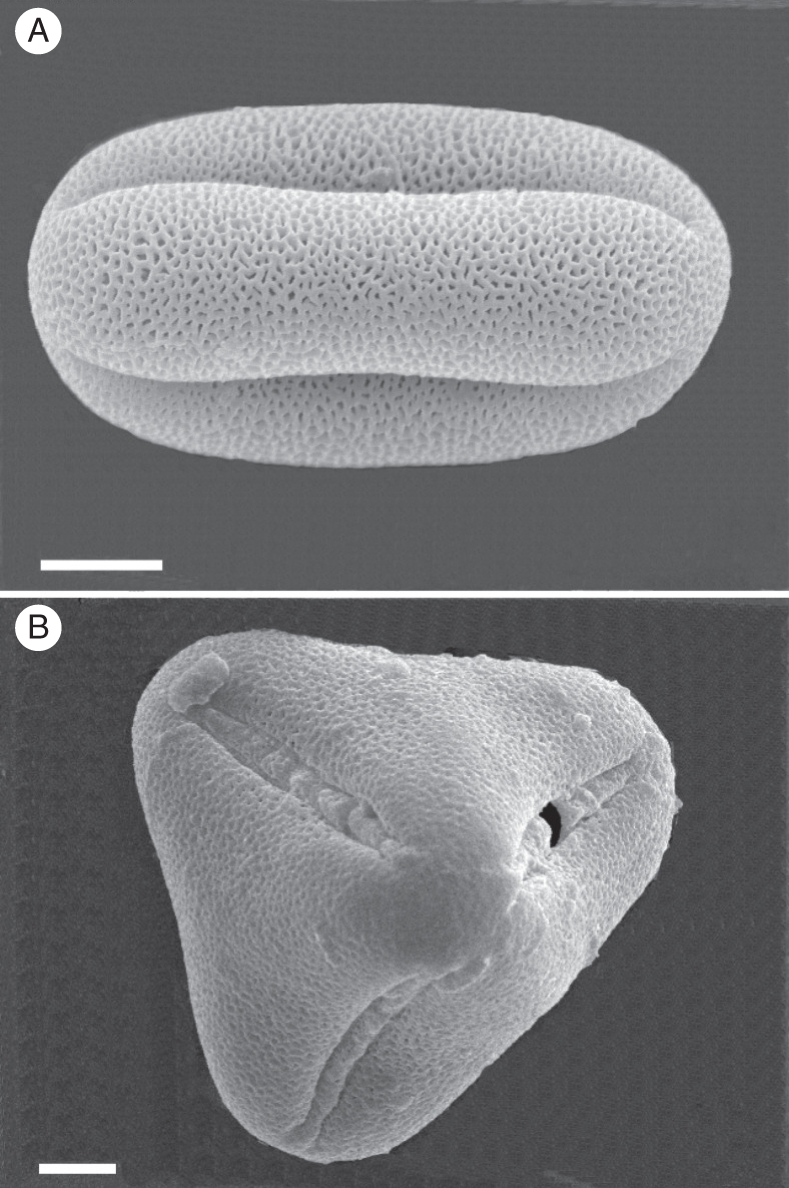
Pollen grains of 2x A. pintoi by SEM: (A) equatorial view of prolate tricolpate reticulate pollen grain; (B) polar view of tetrahedral pollen grain. Scale bars = 10 µm.
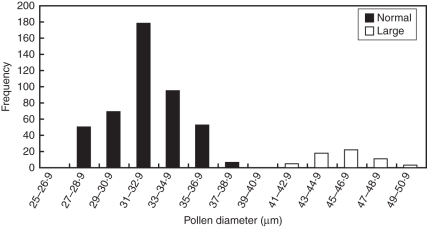
Average pollen grain viability was 93·36 %. Light microscopy analysis of pollen size showed a bimodal distribution (Fig. 6). Based on this result, pollen grains were considered normal when their size ranged between 27·2 and 38·8 µm, and large when the size was above 39·44 µm. With this criterion, the mean size of normal pollen was 32·13 µm, and the frequency was 93·13 %. Large pollen grains with a mean size of 45·7 µm were observed at a frequency of 6·87 %.
Fig. 6.
Distribution of pollen diameter in 2x A. pintoi; normal and large pollen as indicated.
DISCUSSION
In the present study, the nature of a spontaneous triploid of A. pintoi was investigated by analysing somatic chromosomes and meiotic behaviour in both 2x and 3x plants. The analysis of the sporads and pollen grains of diploid plants provided data to reconsider mechanism involved in the origin of polyploid taxa in Arachis.
Evidence of autopolyploidy
The karyotype formula 18m + 2sm found for the diploid A. pintoi with SAT chromosome type 2 agrees with results previously published for the species (Fernández and Krapovickas, 1994). The fact that the karyotype formulae and the SAT chromosomes per haploid complement found in the triploid plants were exactly the same as those observed for the 2x clone is evidence that the 3x has three chromosome sets identical to those observed in the 2x. This observation was confirmed by the physical localization of rDNA by FISH, which showed the same number, size and distribution of the 5S and 18S–26S rDNA loci in each haploid complement of the diploid and triploid plants.
The high frequency of trivalents in all the cells analysed and the presence of up to ten trivalents in diakinesis/metaphase I of the triploid plants indicate a high degree of homology between their three complements. Therefore, the bulk of our evidence demonstrates that the triploid clone of A. pintoi originated by autopolyploidy. Considering the genome nomenclature proposed by Smartt and Stalker (1982) for Arachis species, the genome constitution of this clone would be CCC.
Origin of triploid plants
Polyploidization may be either asexual by somatic chromosome doubling, or sexual through the formation of 2n gametes. As opposed to the early belief of the spontaneous doubling of somatic chromosomes as the main mechanism of polyploid origin, Harlan and De Wet (1975) suggested that polyploids in the angiosperms originated primarily from the union of 2n gametes. More recently, unreduced gametes were considered a dominant process involved in the origin of polyploid plants (Veilleux, 1985; Bretagnolle and Thompson, 1995).
The presence of monads, dyads and triads and large pollen grains constitutes direct evidence that A. pintoi produced 2n gametes. Whereas monads may potentially yield pollen grains with a quadruplicate chromosome number 4n = 40, dyads might give rise to two grains of doubled number 2n = 20, and triads to an unreduced pollen grain 2n = 20, and two pollen grains with reduced chromosome number n = 10. According to Darlington's criterion (Darlington, 1937), a pollen grain is considered unreduced when its size is 1·2–1·3 times larger than n pollen. Therefore, the observations that among the stained pollen the average size of the larger ones was 1·4 times larger than normals also confirmed that unreduced pollen grains were produced by the 2x A. pintoi.
The morphology of normal-sized pollen found here agrees with that described previously for several Arachis species (Pire, 1974; Chaturvedi et al., 1990). Tetrahedral morphology, observed for the large grains, has been reported for Lotus tenuis (Rim and Beuselink, 1996) in which the n pollen grains are globose-prolate, but the 2n pollen grains are tetrahedral. Therefore, the morphological analysis of pollen grains may also support production of unreduced gametes by diploid plants of A. pintoi.
The presence of unreduced gametes in these plants suggests that they have the potential to produce autotriploid plants by unilateral sexual polyploidization via the union of a normal gamete (n) with an unreduced one (2n). Several studies of spontaneous triploids indicate that many of the gametes produced by autotriploids are not functional, because they possess aneuploid, unbalanced chromosome numbers. However, triploids generate small numbers of euploid (x, 2x) gametes and can also produce 3x gametes via nonreduction (Ramsey and Schemske, 1998). Therefore, they may constitute a bridge to produce higher polyploids, like tetraploids and hexaploids by self-fertilization or backcrossing to diploids (Ramanna and Jacobsen, 2003). The finding of unreduced gametes in A. pintoi supports the hypothesis that the allotetraploid A. hypogaea could have also arisen by sexual polyploidization, as suggested by Seijo et al. (2007), and that it may be a general mechanism of polyploid formation in Arachis.
The mechanisms that may give rise to unreduced gametes can be attributed to different failures during the meiotic process. Meiotic irregularities associated with the appearance of 2n gametes are prevalently due to mutations in the genes that regulate chromosome pairing, spindle formation and cytokinesis (Falistoco et al., 1995). Unreduced gametes may result from a process called nuclear restitution; these have been named first-division restitution and second-division restitution, and result from a failure to divide during the first and the second meiotic divisions, respectively (Bretagnolle and Thompson, 1995). This mechanism is considered a potent force in the evolution of some groups of plants, e.g. the Triticeae group (Jauhar, 2007) and Solanum (den Nijs and Peloquin, 1977). Although first-division restitution has been reported for an interspecific triploid hybrid of Arachis (Simpson and Davis, 1983), nuclear restitution was not evident here during the meiotic division of 2x A. pintoi.
Another mechanism that could give rise to unreduced gametes is cytomixis (Falistoco et al., 1995). It has been reported that, as a consequence of this phenomenon, meiocytes that have either fewer or more chromosomes than in normal cells can be formed. Occasionally, either empty meiocytes or meiocytes with a duplicate number of chromosomes have been reported (Bellucci et al., 2003). It is very probable that the meiocytes without chromatin die, but that those with a double number of chromosomes, or almost double, could lead to the formation of 2n gametes. This mechanism is considered by some authors to be of evolutionary importance for plants (Zheng et al., 1987; Falistocco et al., 1995). In the present work, the percentages of cells having cytomictic phenomena (8·77 %), abnormal sporads (8·52 %) and large pollen (6·87 %) were similar in 2x plants, suggesting that cytomixis may be involved in the production of unreduced gametes in A. pintoi.
Nucleolar dominance in autopolyploids
Allopolyploidy induces a wider range of changes in gene expression than does autopolyploidy, suggesting that hybridization has greater effects than genome doubling on gene expression and phenotypic variation (Chen, 2007). Nucleolar dominance is a well-known regulatory phenomenon affecting the active number of NOR loci in allopolyploids and diploid hybrids (Pontes et al., 2004). This silencing mechanism depends on an epigenetic switch that, in concert with promoter methylation and histone modifications, results in a self-reinforcing repression cycle of the rRNA genes (Lawrence et al., 2004). In Triticale (a hybrid between wheat and rye), the wheat NORs are active, and the rye NORs are silenced (Pontes et al., 2004). The generality of this epigenetic mechanism is exemplified in synthetic and natural allopolyploids like Arabidopsis (Madlung et al., 2005) and Glycine (Joly et al., 2004). Evidence of nucleolar dominance in hybrid animals is provided by Drosophila (Durica and Krider, 1977), Xenopus (Pikaard, 1994) and the mule (Kopp et al., 1986).
However, autopolyploids present additional sets of identical genomes, so that the NOR clusters are in extra doses. Considering that nucleolar dominance occurs in allopolyploid species, one can wonder how many clusters are active in the organization of nucleoli in autopolyploids and whether the extra clusters are inactivated. In this work, 3x A. pintoi varied from one to three in the number of active NOR clusters, agreeing with observations for the autopolyploid plant Silene latifolia (Siroki et al., 1999) and the frog Odontophrynus americanus (Ruiz et al., 1980), in which variable numbers of nucleoli have also been found (Beçak and Kobachi, 2004). Therefore, the number of active NORs can vary in both plant and animal autopolyploid species. Those differences in the numbers of chromosomes involved in nucleolar formation observed in autopolyploids cannot be attributed to the main mechanisms of nucleolar dominance proposed in allopolyploids, but could probably be the result of a mechanism of gene dosage compensation.
Conclusions
Karyotype analysis by classical staining, physical localization of chromosome markers by FISH and meiotic analysis revealed that the 3x cytotype of A. pintoi has three identical chromosome complements and demonstrated that this taxa has originated by autopolyploidy. The occurrence of viable unreduced gametes at anthesis of the 2x cytotype suggests that they may have a key role in the polyploidization process of this tropical forage legume. These findings may support unilateral or bilateral sexual polyploidization as a main mechanism in the formation of polyploids within the genus. The occurrence of 2n pollen may allow Arachis breeders to explore new methods for the genetic improvement of peanut and forage species of Arachis. Transfer of desired attributes from variable diploid to polyploid species with a narrow genetic base, like peanut, through sexual polyploidization may be more efficient, in time and product delivery, than other analytical breeding schemes proposed for Arachis so far.
ACKNOWLEDGEMENTS
This research was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) PIP 6265, Secretaría General de Ciencia y Técnica de la Universidad Nacional del Nordeste, PI-47/04, PI-093/07 and Agencia Nacional de Promoción Científica y Tecnológica, PICTO 2007-00099, PICT 2007 No. 1356.
LITERATURE CITED
- Beçak ML, Kobachi LS. Evolution by polyploidy and gene regulation in Anura. Genetics and Molecular Research. 2004;3:195–212. [PubMed] [Google Scholar]
- Bellucci M, Roscini C, Mariani A. Cytomixis in pollen mother cells of Medicago sativa L. Journal of Heredity. 2003;94:512–516. doi: 10.1093/jhered/esg096. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London, Series B, Biological Sciences. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bonavia D. Precerámico peruano, Los Gavilanes, oasis en la historia del hombre. 1982 Corporación Financiera de Desarrollo S.A. COFIDE e Instituto Arqueológico Alemán, Lima, Perú. [Google Scholar]
- Bretagnolle F. Pollen production and spontaneous polyploidization in diploid populations of Anthoxanthum alpinum. Biological Journal of the Linnean Society. 2001;72:241–247. [Google Scholar]
- Bretagnolle F, Thompson JD. Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytologist. 1995;129:1–22. doi: 10.1111/j.1469-8137.1995.tb03005.x. [DOI] [PubMed] [Google Scholar]
- Chaturvedi M, Yunus D, Nair PKK. Cytopalynological studies of Arachis (Leguminosae): cultivated and wild species and their hybrids. Grana. 1990;29:109–117. [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annual Review of Plant Biology. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington CD. Recent advances in cytology. London: J. & A. Churchill; 1937. [Google Scholar]
- Durica DS, Krider HM. Studies on the ribosomal RNA cistrons in interspecific Drosophila hybrids. I. Nucleolar dominance. Developmental biology. 1977;59:62–74. doi: 10.1016/0012-1606(77)90240-8. [DOI] [PubMed] [Google Scholar]
- Erdtman G. Pollen morphology and plant taxonomy: angiosperms. New York, NY: Hafner Publishing Co; 1966. [Google Scholar]
- Falistoco E, Tosti N, Falcinelli M. Cytomixis in pollen mother cells of diploid Dactylis, one of the origins of 2n gametes. Journal of Heredity. 1995;86:448–453. [Google Scholar]
- Fernández A, Krapovickas A. Cromosomas y evolución en Arachis (Leguminosae) Bonplandia. 1994;8:187–220. [Google Scholar]
- Grof B. Forage attributes of the perennial groundnut Arachis pintoi in a tropical savanna environment in Colombia. Proceedings 15th International Grassland Congress, Kyoto. 1985:168–170. Nishi-nasuno, Tochigi-ken, Japan: Science Council of Japan and Japanese Society of Grassland Science. [Google Scholar]
- Harlan JR, De Wet JMJ. On Ö Winge and a prayer: the origins of polyploidy. The Botanical Review. 1975;41:361–390. [Google Scholar]
- Jauhar PP. Meiotic restitution in wheat polyhaploids (amphihaploids): a potent evolutionary force. Journal of Heredity. 2007;98:188–193. doi: 10.1093/jhered/esm011. [DOI] [PubMed] [Google Scholar]
- Jauhar PP, Joppa LR. Chromosome pairing as a tool in genome analysis: merits and limitations. In: Jauhar PP, editor. Methods of genome analysis in plants. Boca Raton, FL: CRC Press; 1996. pp. 9–37. [Google Scholar]
- Jiang J, Gill BS. New 18S–26S ribosomal gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma. 1994;103:179–185. doi: 10.1007/BF00368010. [DOI] [PubMed] [Google Scholar]
- Joly S, Rauscher JT, Sherman-Broyles SL, Brown AHD, Doyle JJ. Evolutionary dynamics and preferential expression of homeologous 18S-5·8–26S, nuclear ribosomal genes in natural and artificial Glycine allopolyploids. Molecular Biology and Evolution. 2004;21:1409–1421. doi: 10.1093/molbev/msh140. [DOI] [PubMed] [Google Scholar]
- Kopp E, Mayr B, Schleger W. Species-specific non-expression in ribosomal RNA genes in a mammalian hybrid, the mule. Chromosoma. 1986;94:346–352. doi: 10.1007/BF00328634. [DOI] [PubMed] [Google Scholar]
- Krapovickas A, Gregory WC. Taxonomía del género Arachis (Leguminosae) Bonplandia. 1994;8:1–186. [Google Scholar]
- Lascano CE, Thomas D. Forage quality and animal selection of Arachis pintoi in association with tropical grasses in the eastern plains of Colombia. Grass & Forage Science. 1988;43:433–439. [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Molecular Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Madlung A, Tyagi AP, Watson B, et al. Genomic change in synthetic Arabidopsis polyploids. The Plant Journal. 2005;41:221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- Moscone EA, Matzke MA, Matzke AJM. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma. 1996;105:231–236. [PubMed] [Google Scholar]
- den Nijs TPM, Peloquin SJ. 2n gametes in potato species and their function in sexual polyploidization. Euphytica. 1977;26:585–600. [Google Scholar]
- Peñaloza APS, Pozzobon MT, Valls JFM. Cytogenetic findings in wild species of Arachis (Leguminosae) Revista Brasileira de Genética. 1996 19(Suplemento): 129. [Google Scholar]
- Pikaard CS. Ribosomal gene promoter domains can function as artificial enhancer of RNA polymerase I transcription, supporting a promoter origin for natural enhancers in Xenopus. Proceedings of the National Academy of Sciences of the USA. 1994;91:464–468. doi: 10.1073/pnas.91.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pire SM. Estudio palinológico de la tribu Hedysareae (Leguminosae) Bonplandia. 1974;12:143–169. [Google Scholar]
- Pontes O, Neves N, Silva M, et al. Chromosomal locus rearrangements are rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proceedings of the National Academy of Sciences of the USA. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt W, Hoen PP, Blackmore S, Nilsson S, Le Thomas A. Glossary of pollen and spore terminology. Review of Palaeobotany and Palynology. 2007;143:1–81. [Google Scholar]
- Ramanna MS, Jacobsen E. Relevance of sexual polyploidization for crop improvement: a review. Euphytica. 2003;133:3–18. [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploidy formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Rim YW, Beuselinck PR. Cytology of 2n pollen formation and pollen morphology in diploid Lotus tenuis (Fabaceae) American Journal of Botany. 1996;83:1057–1062. [Google Scholar]
- Rincón CA, Cuesta PAM, Pérez RB, Lascano CE, Ferguson J. Maní Forrajero Perenne (Arachis pintoi Krapovickas & Gregory): una alternativa para ganaderos y agricultores. Palmira, Instituto Colombiano Agropecuario (ICA) Boletín Técnico. 1992 No. 219. [Google Scholar]
- Robledo G, Seijo JG. Characterization of Arachis D genome by FISH chromosome markers and total genome DNA hybridization. Genetics and Molecular Biology. 2008;31:717–724. [Google Scholar]
- Robledo G, Lavia GI, Seijo JG. Species relations among wild Arachis species with the A genome as revealed by FISH mapping of rDNA loci and heterochromatin detection. Theoretical and Applied Genetics. 2009;118:1295–1307. doi: 10.1007/s00122-009-0981-x. [DOI] [PubMed] [Google Scholar]
- Ruiz IRG, Bonaldo MF, Beçak W. In situ localization of ribosomal genes in a natural triploid of Odontophrynus. Journal of Heredity. 1980;71:55–57. [Google Scholar]
- Schwarzacher T, Ambros P, Schweizer D. Application of Giemsa banding to orchid karyotype analysis. Plant Systematics and Evolution. 1980;134:293–297. [Google Scholar]
- Seijo JG, Lavia GI, Fernández A, et al. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its relatives revealed by double GISH. American Journal of Botany. 2007;94:1963–1971. doi: 10.3732/ajb.94.12.1963. [DOI] [PubMed] [Google Scholar]
- Simpson CE, Davis KS. Meiotic behaviour of a male fertile triploid Arachis L. hybrid. Crop Science. 1983;23:581–584. [Google Scholar]
- Siroki J, Hodurkova J, Negrutiu I, Vyskot B. Functional and structural chromosome analyses in autotetraploid Silene latifolia. Annals of Botany. 1999;84:633–638. [Google Scholar]
- Smartt J, Stalker HT. Speciation and cytogenetics in Arachis. In: Pattee HE, Young CT, editors. Peanut science and technology. Yoakum, TX: American Peanut Research and Education Society; 1982. pp. 21–49. [Google Scholar]
- Stebbins GL. Types of polyploids: their classification and significance. Advances in Genetics. 1947;1:403–429. doi: 10.1016/s0065-2660(08)60490-3. [DOI] [PubMed] [Google Scholar]
- Valls JFM, Simpson CE. New species of Arachis (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia. 2005;14:35–64. [Google Scholar]
- Veilleux R. Diploid and polyploid gametes in crop plants: mechanisms of formation and utilization in plant breeding. Plant Breeding Review. 1985;3:253–288. [Google Scholar]
- Zheng GC, Yang Q, Zheng Y. The relationship between cytomixis, chromosome mutation and karyotype evolution in Lily. Caryologia. 1987;40:243–259. [Google Scholar]