Abstract
Background and Aims
Successful establishment of newly formed polyploid species depends on several interlinked genetic and ecological factors. These include genetic diversity within and among individuals, chromosome behaviour and fertility, novel phenotypes resulting from novel genomic make-up and expression, intercytotypic and interspecific competition, and adaptation to distinct habitats. The allotetraploid rock fern Asplenium majoricum is known from one small population in Valencia, Spain, and several larger populations on the Balearic island of Majorca. In Valencia, it occurs sympatrically with its diploid parents, A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens, and their diploid hybrid A. × protomajoricum. This highly unusual situation allowed the study of polyploid genetic diversity and its relationship to the formation and establishment of nascent polyploid lineages.
Methods
Genetic variation for isozyme and chloroplast DNA markers was determined for A. majoricum and A. × protomajoricum sampled thoroughly from known sites in Majorca and Valencia. Results were compared with variation determined previously for the diploid parent taxa.
Key Results
A highly dynamic system with recurring diploid hybrid and allotetraploid formation was discovered. High diversity in the small Valencian A. majoricum population indicates multiple de novo origins from diverse parental genotypes, but most of these lineages become extinct without becoming established. The populations on Majorca most probably represent colonization(s) from Valencia rather than an in situ origin. Low genetic diversity suggests that this colonization may have occurred only once.
Conclusions
There is a striking contrast in success of establishment of the Majorcan and Valencian populations of A. majoricum. Chance founding of populations in a habitat where neither A. fontanum subsp. fontanum nor A. petrarchae subsp. bivalens occurs appears to have been a key factor enabling the establishment of A. majoricum on Majorca. Successful establishment of this polyploid is probably dependent on geographic isolation from diploid progenitor competition.
Keywords: Asplenium majoricum, Asplenium × protomajoricum, A. fontanum subsp. fontanum, A. petrarchae subsp. bivalens, allopolyploid, multiple origins, hybridization, colonization, Balearic islands, polyploid speciation
INTRODUCTION
Polyploidy is an important process in the evolution of new plant species, with virtually all plants, including ferns, thought to have undergone at least one round of polyploidization in their evolutionary history (Soltis and Soltis, 2009). It has been estimated that polyploidy is involved in some 15 % of speciation events in angiosperms and 31 % in ferns (Wood et al., 2009). The genetic and ecological factors responsible for the evolutionary success of polyploids are the subject of ongoing debate and research (Soltis and Soltis, 2000; Ramsey and Schemske, 2002; Coyne and Orr, 2004; Guggisberg et al., 2006; Dubcovsky and Dvorak, 2007; Hegarty and Hiscock, 2008; Meimberg et al., 2009), with different studies emphasizing different factors. Increased within-individual genetic variation is provided by fixed heterozygosity (Soltis and Soltis, 2000; Soltis et al., 2004; Meimberg et al., 2009), which may act as a buffer against deleterious recessive alleles and hence facilitate the transition to inbreeding observed in many polyploids (Vogel et al., 1999a; Guggisberg et al., 2006; Barringer, 2007; Husband et al., 2008). Set against this increased within-individual variation is the reduced population-level diversity that results from the genetic bottleneck associated with polyploid speciation (Coyne and Orr, 2004; Meimberg et al., 2009). This genetic bottleneck will be less severe when a new polyploid species arises from multiple independent polyploidization events. Multiple origins have been demonstrated in the majority of polyploid species studied (Soltis and Soltis, 1999; Soltis et al., 2004) and this may lead to parallel speciation processes (Perrie et al., 2010). Studies on bread wheat (Triticum aestivum) and Aegilops triuncualis have suggested that the increased genetic diversity provided by multiple origins directly contributes to ecological tolerance and evolutionary success (Dubcovsky and Dvorak, 2007; Meimberg et al., 2009).
Evolutionary success of novel polyploids is contingent not only on genetic and ecological factors but also on the dynamics of their establishment (Whitton, 2004). In the initial generations following polyploid species formation, polyploid success may be the exception rather than the norm. Novel tetraploid taxa, arising sporadically de novo in populations of their parents, may be expected to have lower reproductive success due to the effect of minority cytotype exclusion: the majority of offspring of tetraploid individuals will be sterile triploids, produced by back-hybridization with a diploid parent (Levin, 1975). Many newly formed polyploids will thus constitute evolutionary dead-ends. Establishment and spread of polyploid populations can be expected to require reproductive isolation from parental taxa (Coyne and Orr, 2004; Whitton, 2004), for example by geographical isolation or by a change in breeding system to reduce the tendency for back-crossing.
In the current study, these dynamics were investigated in the allotetraploid (2n = 144) rock fern Asplenium majoricum. Asplenium majoricum was first described from Majorca (Litardière 1911), and originally thought to be endemic to the island, where it grows predominantly on limestone walls of olive terraces at altitudes of 100–350 m (Sleep, 1967; Jacquotot and Orell, 1968). Later, its presence at two sites in Valencia in mainland Spain was confirmed by Pérez Carro and Fernández Areces (1992). Cytological studies identified the diploid (2n = 72) parents of A. majoricum as A. fontanum subsp. fontanum (Sleep, 1967) and A. petrarchae subsp. bivalens (Lovis and Reichstein, 1969). Both these taxa are calcicoles, growing in fissures and crevices of limestone rock outcrops. Asplenium petrarchae subsp. bivalens, which grows up to 700 m altitude, is more thermophilic than A. fontanum subsp. fontanum, which grows up to 2100 m (Ibars et al., 1999; Hunt et al., 2009). Only a few localities provide the opportunity for the formation of hybrids between these two species: they rarely grow sympatrically because of their different ecological preferences, and because A. petrarchae subsp. bivalens is a rare fern with a narrow distribution (Ibars et al., 1999; Hunt et al., 2009). Their diploid hybrid, A. × protomajoricum, has been reported from five sites in Valencia and Alicante (Pangua et al., 1992; Pérez Carro and Fernández Areces, 1992; Herrero Nieto and Prada Moral, 1997). Asplenium × protomajoricum is largely sterile, with most spores abortive. Production of diplospores occurs at low frequency, however, suggesting a mechanism for the origin of the allotetraploid A. majoricum (Pangua et al., 1992; Pérez Carro and Fernández Areces, 1992).
Several factors make this species and its progenitors an ideal system in which to study the dynamics of incipient polyploid speciation. The distributions and reticulate relationships of these and other Asplenium species and hybrids in southwestern Europe are exceptionally well studied, with detailed accounts by Reichstein (1981) and Sleep (1983). The occurrence together at a single site of both diploid parents, a diploid hybrid and a derived allotetraploid is highly unusual among polyploids (Levin, 2002). We previously analysed the phylogeography of the two parental species by studying the geographic differentiation of chloroplast haplotype and isozyme allele frequencies (Hunt et al., 2009). This knowledge potentially enables determination of the likely geographic origin of A. majoricum populations. We are hence able to address the question of whether A. majoricum arose in situ on Majorca, or colonized the island following an origin on the mainland Iberian peninsula. The two parental species do not co-occur on Majorca today. Asplenium petrarchae subsp. bivalens is found at a small number of sites on Majorca (Bennert et al., 1990; Hunt et al., 2009). Asplenium fontanum subsp. fontanum was recorded there in 1917 but has not been found since. However, the presence of triploid hybrids, A. × reichsteinii, with parentage A. majoricum × A. fontanum subsp. fontanum, suggests that the latter taxon may persist on Majorca but is very rare (Bennert et al., 1987).
In the current study, the following questions were addressed. Do allozyme and chloroplast DNA markers enable the tracing of inheritance of alleles from the diploid parents A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens through the diploid hybrid A. × protomajoricum into the allotetraploid A. majoricum? Does the genetic evidence corroborate previous morphological and cytological data on the reticulate evolutionary relationships between these taxa? How many events gave rise to the polyploids that founded the current populations of A. majoricum, and where were these events located? What factors appear to be significant in enabling the establishment of these polyploid lineages, comparing the Valencian and Majorcan allotetraploid populations?
MATERIALS AND METHODS
Plant collection
All recorded sites for Asplenium × protomajoricum and A. majoricum (Pangua et al., 1992; Pérez Carro and Fernández Areces, 1992; Herrero Nieto and Prada Moral, 1997) in mainland Spain were searched either once or twice. Intensive sampling was carried out at these sites of putative hybrids and allotetraploids and their diploid parents A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. The diploid hybrid A. × protomajoricum is morphologically similar to the tetraploid A. majoricum (Pangua et al., 1992; Pérez Carro and Fernández Areces, 1992). Pinnae and rhizome scales may be somewhat smaller in the former, but these are not reliable diagnostic characters. The chromosome number represents the key difference between the two taxa, but performing chromosome counts on large numbers of plants is impractical. Spore morphology was therefore used to discriminate between A. × protomajoricum and A. majoricum. Sporangia of A. × protomajoricum are mostly abortive with deformed spores and irregular dark deposits (Pangua et al., 1992; Pérez Carro and Fernández Areces, 1992). Diplospores, i.e. viable unreduced spores which appear large, regular in shape and pale brown, are often present, but are in a minority. This is characteristic of most fern hybrids, which are typically highly sterile; fertile hybrids are very rare (in contrast to flowering plants; Reichstein, 1981; Moran, 1982; Wagner et al., 1985). The strongly bimodal distribution of diplospore frequency therefore makes this a reasonable character to determine whether a plant appearing intermediate morphologically between A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens is A. × protomajoricum or A. majoricum.
The numbers of plants of each taxon collected from each site are given in Table 1. A total of 26 individuals of A. × protomajoricum were found at three sites: three individuals from Barranc de las Cuevas (BDLC), near Moixent, 11 individuals from a disused quarry near Simat de la Valldigna (SDLV) and 12 individuals from La Gola (LG), near Font Nova. Asplenium majoricum (29 individuals) was found in Valencia only at Barranc de las Cuevas. Four populations of A. majoricum on Majorca were also sampled, for a total of 350 plants (Table 1). Three of these populations (MAJ-1, MAJ-2 and MAJ-3) were collected from limestone walls bounding olive terraces near Sóller, the habitat from which this species was originally described (Sleep, 1983). The fourth population (MAJ-4) was collected from natural limestone outcrops near Caimari in the northeast of the island.
Table 1.
Details of collection sites for A. majoricum and A. × protomajoricum for this study. Samples of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens collected from sites in Valencia where these taxa grow sympatrically with A. × protomajoricum and/or A. majoricum are also shown. These latter samples were analysed previously in Hunt et al. (2009).
| Number of samples |
||||||||
|---|---|---|---|---|---|---|---|---|
| Region | Site | Latitude | Longitude | Code and collection year | A. fontanum subsp. fontanum | A. petrarchae subsp. bivalens | A. × protomajoricum | A. majoricum |
| Valencia | Barranc de las Cuevas | N38°51'13'' | W000°43'03'' | BDLC-99 | 11 | 20 | 0 | 9 |
| BDLC-02 | 7 | 20 | 3 | 20 | ||||
| Valencia | Simat de la Valldigna | N39°02'29'' | W000°20'30'' | SDLV-99 | 44 | 94 | 3 | 0 |
| SDLV-02 | 60 | 52 | 8 | 0 | ||||
| Valencia | La Gola | N39°01'38'' | W000°16'28'' | LG-02 | 11 | 22 | 12 | 0 |
| Majorca | Ses Tres Creus | N39°45'42'' | E002°43'00“ | MAJ-1 | 0 | 0 | 0 | 120 |
| Majorca | Ses Tres Creus | N39°45'40'' | E002°43'1'' | MAJ-2 | 0 | 0 | 0 | 126 |
| Majorca | Es Verger, Biniarix | N39°46'10'' | E002°44'25'' | MAJ-3 | 0 | 0 | 0 | 88 |
| Majorca | Torrent d'es Horts, Caimari | N39°46'42'' | E002'53'55“ | MAJ-4 | 0 | 7 | 0 | 16 |
| Summary of additional samples from sites without hybrids or allotetraploids* | ||||||||
| Majorca | 0 | 57 (2) | 0 | 0 | ||||
| Valencia | 342 (5) | 349 (6) | 0 | 0 | ||||
| Aragon | 400 (11) | 0 | 0 | 0 | ||||
| Catalunya | 67 (3) | 0 | 0 | 0 | ||||
| Pyrenees | 449 (14) | 0 | 0 | 0 | ||||
| Alpes-Maritimes | 423 (14) | 0 | 0 | 0 | ||||
| Switzerland | 84 (3) | 0 | 0 | 0 | ||||
* Numbers in parentheses indicate the number of sites in each region.
Additional isozyme data covering the entire distribution ranges of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens (Hunt et al., 2009) were used in conjunction with the new data reported here on A. × protomajoricum and A. majoricum and integrated in analyses as detailed below.
Allozymes
Allozyme variation was determined for all individuals sampled. Proteins were extracted in a Tris–HCl (pH 7·5) buffer (Soltis et al., 1983), and fractionated on 13·7 % hydrolysed potato starch gels following established protocols (Wendel and Weeden, 1989; Vogel et al., 1999a). Five enzymes were resolved on the lithium borate gel and electrode buffer system 8 of Soltis et al. (1983): acid phosphatase (ACP; EC 3·1·3·2), aspartate aminotransferase (AAT; EC 2·6·1·1), phosphoglucose isomerase (PGI; EC 5·3·1·9), phosphoglucomutase (PGM; EC 5·4·2·2) and triose-phosphate isomerase (TPI; EC 5·3·1·1). Six enzymes were resolved using a morpholine citrate (pH 7·0–7·4) gel and electrode buffer system (Wendel and Weeden, 1989): 6-phosphogluconate dehydrogenase (6-PGD; EC 1·1·1·44), hexokinase (HEX; EC, 2·7·1·1), isocitrate dehydrogenase (IDH; EC 1·1·1·42), malate dehydrogenase (MDH; EC 1·1·1·37), PGM, shikimate dehydrogenase (SKDH; EC 1·1·1·25) and UTP-glucose pyrophosphorylase (UGPP; EC 2·7·9). Allozyme banding systems were interpreted using known enzyme sub-structuring (Kephart, 1990), taking into account the results of previous Asplenium allozyme studies (Vogel et al., 1999a; Suter et al., 2000) and our previous results on the two diploid parent taxa, A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens (Hunt et al., 2009). For enzymes with multiple loci, the most anodally migrating isozyme was designated locus-1.
Ancestral identity of alleles was established by running samples of A. × protomajoricum and A. majoricum together with their diploid parents A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. As described previously (Hunt et al., 2009) alleles in these two taxa were labelled according to their relative mobility to a high-frequency ‘100’ allele in each taxon. All alleles known in these taxa from across their entire respective distribution ranges are shown in Table S1 (Supplementary Data, available online). At each locus, most phenotypes in A. × protomajoricum and A. majoricum could be interpreted as a compound allele comprising one allele from A. fontanum subsp. fontanum and one from A. petrarchae subsp. bivalens, or in some cases a null allele from one or other parent. Each such compound allele was allocated a letter code and plants were scored accordingly for these as haploid data for A. × protomajoricum and diploid for A. majoricum. Codes were allocated such that the same letter code in A. × protomajoricum and A. majoricum represents the same phenotype. Unbalanced banding patterns, i.e. atypical heterozygous patterns in which bands stain with unequal intensities, indicating gene dosage effects, were used to infer heterozygosity for two different compound alleles in A. majoricum.
These data were used to estimate genetic diversity statistics and population structure and infer multilocus genotypes (MLGs). Some individuals could not be assigned a certain MLG due to missing data. These were allocated a putative MLG, designated with a subscript letter. Following preliminary analyses which indicated negligible genetic differentiation between Majorcan populations of A. majoricum (data not shown), plants were treated as two populations, from Valencia (Barranc de las Cuevas) and Majorca (merged across all four sampled sites) for all subsequent analyses. The genetic diversity statistics percentage of polymorphic loci by the 95 and 99 % criteria (P95 and P99), number of alleles (A), number of alleles per polymorphic locus (Ap), expected heterozygosity (He) and observed heterozygosity (Ho) were estimated in GDA ver. 1·1 (Lewis and Zaykin, 2001). The effective number of alleles (Ae) was calculated in PopGene ver. 1·32 (Yeh et al., 1999). Allelic richness (R) with correction for the smallest regional sample size (n = 26) through rarefaction (El Mousadik and Petit, 1996; Petit et al., 1998) was calculated using Fstat v. 2·9·3 (Goudet, 1995).
Population structure in A. majoricum was analysed in GDA to give estimates of Wright's (1978) F-statistics (FIS, FIT and FST) and associated 95 % confidence intervals (CIs; obtained by bootstrapping with 9999 replicates). MLG diversity [1 – Σxi2, a statistic comparable with the chloroplast haplotype diversity statistic used by Trewick et al. (2002), where xi is the frequency of the ith MLG] was calculated in Microsoft Excel.
Neighbor–Joining phenograms of relationships between MLGs in A. × protomajoricum and A. majoricum were constructed in PowerMarker ver. 3·25 (Liu and Muse, 2005) based on matrices using the genetic distance measure of Nei et al. (1983). Analyses using a range of alternative distance measures were tested and gave tree topologies with only very minor variations (data not shown). The genotypes in A. × protomajoricum were duplicated so that the whole data set could be treated as diploid in this analysis. Trees were manipulated in Dendroscope ver. 2·2 (Huson et al., 2007).
To enable evaluation of hypotheses regarding the geographical origins of A. majoricum, the frequencies of the alleles in the allotetraploid that had been identified as originating from A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens were compared with their frequencies in these diploid taxa. The raw data previously described in Hunt et al. (2009) were used in this analysis, comprising 1898 plants of A. fontanum subsp. fontanum from 53 populations and 621 plants of A. petrarchae subsp. bivalens from 12 populations. A summary of the geographic origins of these samples is given in Table 1. Allele frequencies in samples of A. fontanum subsp. fontanum, A. petrarchae subsp. bivalens and A. majoricum were calculated in PowerMarker ver. 3·25 (Liu and Muse, 2005).
Chloroplast DNA
As in most flowering plants, chloroplast DNA is inherited maternally in Asplenium (Vogel et al., 1998a). Restriction fragment analysis of the the trnLUAA–trnFGAA region, including the trnL intron and the intergenic spacer (IGS) between trnL and trnF (henceforth abbreviated as ‘trnL-F’) was used to determine the maternal parent of A. × protomajoricum and A. majoricum. DNA extraction followed the procedure of Schneider et al. (2004). Amplifications using the primers Fern-1 (Trewick et al., 2002) and F (Taberlet et al., 1991) were performed according to Trewick et al. (2002) and Schneider et al. (2004). Initial sequence analysis of this region in A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens found considerable differentiation between these taxa (91 % sequence identity; Supplementary Data Fig. S1, available online). This differentiation included a consistent interspecific polymorphism for an EcoRV restriction site that was present in A. fontanum subsp. fontanum and absent in A. petrarchae subsp. bivalens (Fig. S1). This site showed no intraspecific variation within either taxon, from plants sampled from across the entire respective ranges of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. The trnL-F region in 20 plants of A. × protomajoricum was then amplified (eight from population SDLV; 12 from LG) and 30 plants of A. majoricum (one from the Valencian population BDLC, ten from the Majorcan population MAJ-1, nine from MAJ-2 and ten from MAJ-3). Restriction digests were performed in 10 µL volumes containing 5 µL of PCR product, 1 µL of EcoRV (Promega) and 1 µL of Buffer D (Promega). Digestion products were electrophoresed on agarose gels and visualized under UV light with ethidium bromide. Samples of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens were used as standards for comparison with the hybrid and polyploid samples.
Following results of this analysis, which indicated that A. petrarchae subsp. bivalens was the maternal parent of all A. × protomajoricum and A. majoricum samples (see Results), the trnL-F region in 24 samples of A. × protomajoricum (ten from population SDLV; three from LG; and 11 from BDLC) and 22 of A. majoricum (11 from the Valencian population BDLC; and 11 from among the Majorcan populations MAJ-1 to MAJ-4) was sequenced to enable comparison with the A. petrarchae subsp. bivalens trnL-F haplotype data set obtained previously (Hunt et al., 2009). PCR products were sequenced for both strands using BigDye Terminator Kits (v. 1·1 Applied Biosystems, ABI) and an ABI 3730 capillary DNA sequencer. Sequences were assembled using SeqMan ver. 3·56 (DNASTAR, Inc.), and aligned together with our previous trnL-F haplotype data set for A. petrarchae subsp. bivalens (Hunt et al., 2009), using MegAlign ver. 3·14 (DNASTAR, Inc.), and checked manually. In this study all variable sites including indels were retained in the analysis. Most indel variation resulted from length changes in a poly(C) microsatellite. We recognize that the relationships between haplotypes differing in the number of repeat units may be ambiguous because of uncertainty over whether a stepwise mutation model is appropriate. Novel sequences have been submitted to GenBank (accession numbers GU017743 – GU017745; Table 2). Haplotype assignation and network construction were carried out in Arlequin ver. 3·0 (Excoffier et al., 2005).
Table 2.
Definition of trnL-F haplotypes in A. petrarchae subsp. bivalens (PB), A. × protomajoricum (PM) and A. majoricum (M), and associated GenBank accession numbers
| Alignment position |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype number* | GenBank accession no. | 29 | 156 | 309 | 753 | 754 | 755 | 779 | 804 | 834 | Present in taxa |
| 1 | FJ456852 | C | – | T | – | – | – | G | G | T | PB |
| 2 | FJ456853 | A | T | T | – | – | – | G | A | T | PB, PM, M |
| 3 | FJ456854 | A | T | C | C | – | – | G | G | T | PB |
| 4 | FJ456855 | A | T | T | C | – | – | G | G | T | PB, PM |
| 5 | FJ456856 | A | T | T | C | C | – | G | G | T | PB |
| 6 | FJ456857 | A | T | T | – | – | – | T | G | T | PB |
| 7 | GU017743 | A | T | T | C | C | C | G | G | T | M |
| 8 | GU017744 | A | T | T | C | – | – | G | G | G | PM |
| 9 | GU017745 | A | T | T | – | – | – | G | G | T | PM |
* These numbers are also used in Fig. 4.
RESULTS
Identification of hybrid and allotetraploid individuals
Based on frond and sporangial morphology, 26 individuals were identified as diploid hybrids (as detailed above). A total of 379 individuals were identified as A. majoricum. The morphological characteristics that were used to discriminate between the hybrid and allotetraploid were those described in Pangua et al. (1992) and Pérez Carro and Fernández Areces (1992). Chromosome counts on type specimens from these precise localities by these authors have previously established the relationship between ploidal level and sporangial morphology, which is also typical for other homosporous ferns (Moran, 1982; Wagner et al., 1985). Morphological discrimination of hybrid individuals from A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens was supported by their characteristic isozyme banding patterns (described below).
Allozymes
Thirteen loci were resolved and scored in A. × protomajoricum and A. majoricum. Twelve of these were also scored in both parental species, A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens (Hunt et al., 2009). One locus, MDH-1, was also scored in A. petrarchae subsp. bivalens but not in A. fontanum subsp. fontanum, where it was not reliably resolved. Each distinct banding pattern at each locus was allocated a letter code. Comparison of the banding patterns with the sets of alleles in A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens enabled interpretation of observed phenotypes in terms of additive banding patterns in the hybrids and allotetraploids from the alleles donated by the two parental taxa (Supplementary Data Table S2, available online).
In A. × protomajoricum, seven loci (ACP, HEX, MDH-1, MDH-2, PGM-2, TPI-1 and TPI-2) showed a single phenotype across the 26 plants analysed. The number of phenotypes at polymorphic loci ranged from two (UGPP and IDH) to five (6-PGD and AAT-1). Most of these phenotypes clearly represented additive banding patterns between alleles identified in A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. Exceptions were phenotype E in 6-PGD, phenotype B in SKDH and the four phenotypes in MDH-1. In the first two of these cases, the phenotype in A. × protomajoricum apparently lacks an allele from one of the two parental species. This could indicate a loss-of-function mutation in one of the two genomes of the hybrid, resulting in a null allele. Alternatively, sampling of the parental taxa could have missed, by chance, an allele which has similar mobility to that contributed by the other diploid parent, such that the hybrid phenotype appears as a single band. The locus MDH-1 was not scored in A. fontanum subsp. fontanum, because co-migration with MDH-2 prevented consistent resolution of alleles. However, it was observed that some populations of A. fontanum subsp. fontanum showed an allele that could unambiguously be attributed to MDH-1. This allele had the same mobility as the 100 allele in A. petrarchae subsp. bivalens.
In A. majoricum, five loci (ACP, HEX, MDH-2, TPI-1 and TPI-2) were monomorphic for a single phenotype. The number of phenotypes at polymorphic loci ranged from two (6-PGD, IDH, PGI-2 and UGPP) to four (AAT-1 and MDH-1). In most cases, the phenotypes observed in A. majoricum were also present in A. × protomajoricum. At the loci MDH-1, PGM-2, SKDH and UGPP, phenotypes specific to A. majoricum were observed that could result from the presence of a null allele in one or other homeologous genome. At MDH-1 and AAT-1, one phenotype (D and F, respectively) was not found in A. × protomajoricum, but clearly combines alleles present in A. petrarchae subsp. bivalens and A. fontanum subsp. fontanum. A second phenotype at AAT-1 (G) contains an allele (84) which we infer to be derived from A. fontanum subsp. fontanum, but which was not found in any of the populations of this taxon we analysed previously (Hunt et al., 2009; see Supplementary Data Table S1).
In the tetraploid A. majoricum, most individuals exhibited balanced banding patterns at most loci, which indicated they were homozygous for one particular compound allele. At the loci AAT-1 and IDH, a small number of plants from the Valencian population showed unbalanced banding patterns (Supplementary Data Table S2) that are consistent with heterozygosity for two different compound alleles, suggesting that they arose from crosses between gametophytes of different genotypes.
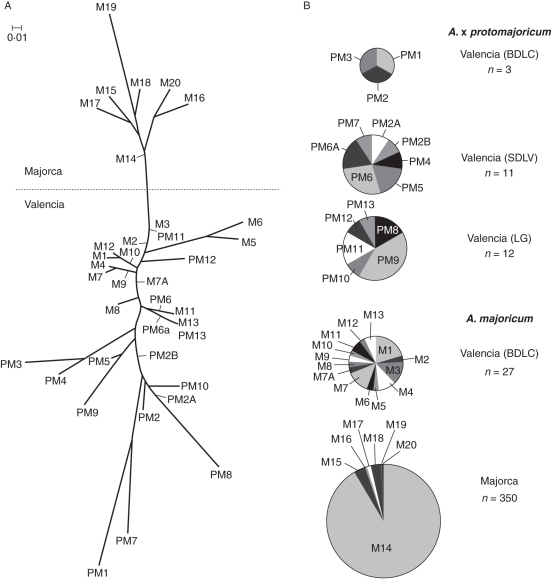
The phenotypes across the 13 loci occurred in combinations to yield 13 MLGs in A. × protomajoricum and 20 in A. majoricum. In A. majoricum, none of the MLGs was shared between the Majorcan and Valencian populations: seven were specific to the former, and 13 to the latter. In the Valencian population, one MLG was shared by plants collected in 1999 and 2002. Two of these individuals were excluded from further analysis, as these might have represented re-sampling of the same plant. Otherwise, there were no MLGs in common between the nine A. majoricum plants collected in 1999 and the 20 collected in 2002. MLGs for the samples are shown in Supplementary Data Table S3. The genetic distances between the MLGs are given in Supplementary Data Table S4. Neighbor–Joining phenograms of the MLGs are shown in Fig. 1A, and reveal that the Majorcan MLGs form a cluster distinct from those in the Valencian population. The most common MLG in Majorca (M14, 92 % frequency; Fig 1B) occupies a central position in this cluster, and has the shortest genetic distance to the group of MLGs from Valencian A. majoricum. The Valencian MLGs form a group on the tree which also contains some of the MLGs from A. × protomajoricum. Two of the MLGs in A. majoricum (M3 and M13) were identical to one in A. × protomajoricum (PM11 and PM13, respectively), although neither of the latter were from the Barranc de las Cuevas site where the A. majoricum population was found. The tree also shows a group of A. × protomajoricum MLGs that are more genetically distinct from any of the A. majoricum types. Differentiation (FST) between the Majorcan and Valencian populations of A. majoricum was estimated at 0·857, although the wide 95 % CI (0·055–0·970) makes it difficult to gauge the extent of differentiation.
Fig. 1.
(A) Neighbor–Joining phenogram based on the Nei (1983) genetic distance matrix, showing relationships between multilocus genotypes in A. × protomajoricum (PM) and A. majoricum (M). Narrow lines indicate where labels relate to internal nodes. (B) Frequency of MLGs in A. × protomajoricum and A. majoricum by sampled population.
Table 3 compares the diversity statistics in the Majorcan and Valencian populations of A. majoricum, based on both MLG and compound-allele distributions. Although the Valencian population is less than a tenth of the size of the Majorcan population, it shows higher diversity as assessed by the number of polymorphic loci, effective number of alleles, expected and observed heterozygosity, and MLG richness and diversity. The Majorcan population has higher allelic richness, but lower allelic richness when sample size is taken into account using rarefaction. Inbreeding (FIS) estimates were 0·679 (95 % CI = 0·306–1·000) for the Valencian population and 1·000 (95 % CI = 1·000–1·000) for the Majorcan population.
Table 3.
Isozyme genetic diversity in A. majoricum for the populations from Valencia (BDLC) and Majorca
| Population | n | No. of MLGs | MLG diversity = 1 – Σi2 | P99 | A | Ap99 | Ae | He | Ho |
|---|---|---|---|---|---|---|---|---|---|
| BDLC | 27 | 13 | 0·893 | 0·231 | 1·385 | 2·667 | 1·237 | 0·097 | 0·032 |
| Majorca | 350 | 7 | 0·152 | 0·154 | 1·538 | 3·500 | 1·014 | 0·013 | 0·000 |
The genetic diversity statistics are based on means across 13 loci.
n, number of individuals; MLG, multilocus genotype; P, proportion of polymorphic loci; A, number of alleles per locus; Ap, number of polymorphic alleles per locus; Ae, effective number of alleles per locus; He, expected heterozygosity; Ho, observed heterozygosity. 99 % criterion used for determining polymorphic loci. Diversity statistic estimates correct to 3 decimal places.
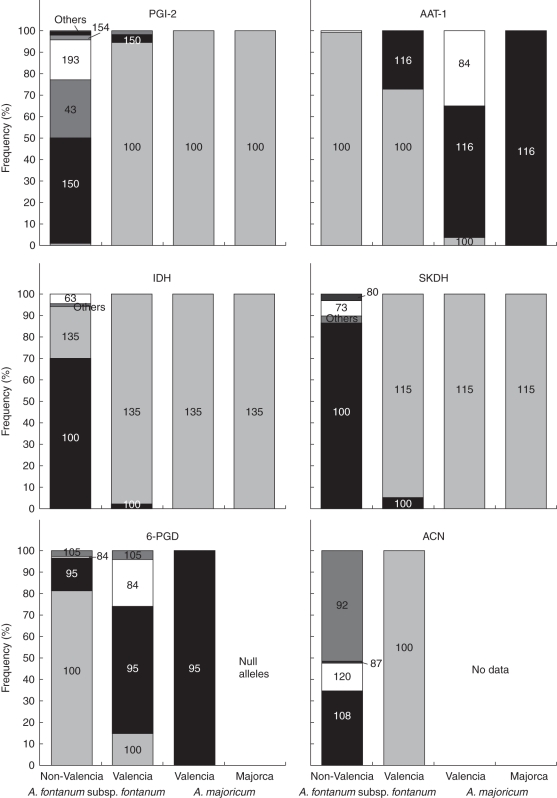
The geographic distribution of alleles in A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens is informative in assessing the origin(s) of populations of A. majoricum. In A. fontanum subsp. fontanum, the Valencian populations are strongly genetically differentiated from those elsewhere in the range by distinct allele frequencies at a number of loci [PGI-2, AAT-1, IDH, SKDH, 6-PGD and ACN (Hunt, 2004; Hunt et al., 2009)]. Figure 2 shows allele frequencies at these loci for Valencian and non-Valencian populations of A. fontanum subsp. fontanum. The frequencies of the alleles in Valencian and Majorcan A. majoricum derived from A. fontanum subsp. fontanum are also shown in Fig. 2. One of these loci, ACN, was not scored in A. majoricum, while 6-PGD appeared to be represented by a null allele in Majorcan populations of A. majoricum. At the remaining four loci, the data demonstrate that almost all of the alleles in A. majoricum which are derived from A. fontanum subsp. fontanum are found in the Valencian populations of A. fontanum subsp. fontanum. The exception is the AAT-1 84 allele that we inferred was derived from A. fontanum subsp. fontanum (discussed below). The data from A. fontanum subsp. fontanum are therefore consistent with a Valencian origin for both the Valencian and Majorcan populations of A. majoricum. However, because A. fontanum subsp. fontanum appears to be extinct or near-extinct on Majorca, it is not known whether these same alleles are or were also found in Majorcan populations of this taxon. These data do not, therefore, exclude the possibility that Majorcan populations of A. majoricum originated in situ on the island. Several of the alleles in A. majoricum donated by A. fontanum subsp. fontanum are absent from or extremely rare in populations of the latter taxon outside Valencia (i.e. in its range in mainland Europe from Aragon in eastern Spain through Catalunya, the Pyrenees, southeastern France and the Swiss Jura). This makes it highly unlikely that any of these populations of A. fontanum subsp. fontanum contributed to the origin of A. majoricum, although in any case there was no reason why this might have been expected.
Fig. 2.
Frequencies of alleles in A. majoricum derived from A. fontanum subsp. fontanum, and their frequencies in populations of this diploid taxon, compared for Valencian and non-Valencian populations of A. fontanum subsp. fontanum and for Valencian and Majorcan populations of A. majoricum. Numbers on the bars indicate allele names, as detailed in Supplementary Data Table S2.
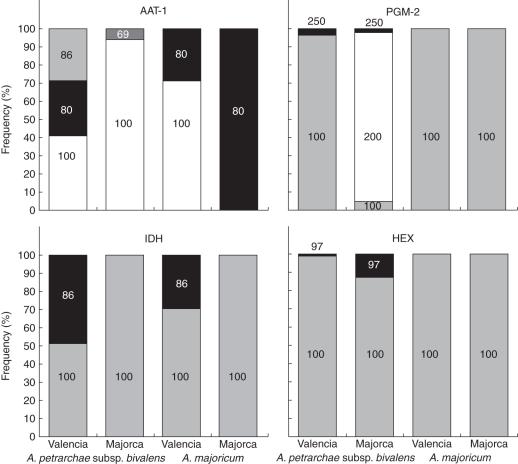
In A. petrarchae subsp. bivalens, populations on Majorca are genetically differentiated from those in Valencia; loci implicated in this differentiation are AAT-1, HEX, IDH-1 and PGM-2 (Hunt, 2004; Hunt et al., 2009). Bar plots of allele frequencies at these loci are shown in Fig. 3. All other loci had a homogeneous (and near-monomorphic) distribution of allele frequencies in both taxa.
Fig. 3.
Frequencies of alleles in A. majoricum derived from A. petrarchae subsp. bivalens, and their frequencies in populations of this diploid taxon, compared for Valencian and Majorcan populations. Numbers on the bars indicate allele names, as detailed in Supplementary Data Table S2.
Two of the above loci, AAT-1 and PGM-2, are informative with regard to a possible Majorcan origin of the allotetraploid. At AAT-1, the Majorcan A. majoricum populations are monomorphic for the 80 allele derived from A. petrarchae subsp. bivalens. This allele is present at 30 % frequency in Valencian populations of A. petrarchae subsp. bivalens, but absent from those in Majorca. At PGM-2, the 100 allele is at very high (97 %) frequency in A. petrarchae subsp. bivalens in Valencia, but very low (5 %) frequency in Majorca, which is dominated by the 200 allele. Valencian and Majorcan populations of A. majoricum are both monomorphic for the 100 allele. At the remaining loci, IDH is polymorphic for the 100 and 86 alleles in both A. petrarchae subsp. bivalens and A. majoricum in Valencia, but monomorphic for the 100 allele in both taxa in Majorca. HEX shows a higher frequency of the 97 allele in A. petrarchae subsp. bivalens in Majorca than in Valencia, but this allele is still uncommon relative to the 100 allele in both populations, and thus the monomorphism of A. majoricum populations in both locations for this allele is unremarkable.
Chloroplast DNA
EcoRV digests of the approx. 900 bp trnL-F amplicons gave products approx. 900 bp in size for A. petrarchae subsp. bivalens and approx. 450 bp for A. fontanum subsp. fontanum. All 29 individuals of A. × protomajoricum and 20 of A. majoricum analysed by the same method yielded approx. 900 bp products, indicating that A. petrarchae subsp. bivalens was the maternal parent in all cases for both the diploid hybrid and its derived allotetraploid.
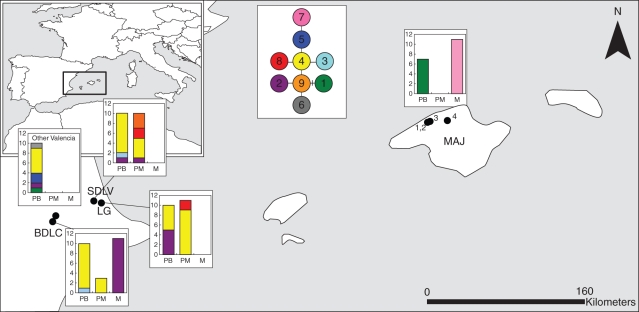
DNA sequencing of the approx. 900 bp trnL-F region identified a total of nine haplotypes among the A. petrarchae subsp. bivalens, A. × protomajoricum and A. majoricum samples (Table 2). A number of these haplotypes differ from one another only in the number of repeat units in a mononucleotide motif. Although such runs are highly susceptible to homoplasy, these sites were included in the analysis because we found that excluding them gave the impression of sequence identity between A. majoricum and the other two taxa, which we judged as potentially more misleading than the consequences of homoplasy. The minimum spanning network of relationships between the haplotypes and their geographical distribution are shown in Fig. 4. Six haplotypes were found in A. petrarchae subsp. bivalens, of which four (1, 3, 5 and 6) were restricted to this diploid progenitor. In this species, six haplotypes were detected among the 40 Valencian mainland samples (1–6), with haplotype 4 the most common (68 %), while the seven Majorcan samples were invariant for haplotype 1, as previously described (Hunt et al., 2009). Four haplotypes (2, 4, 8 and 9) were present in the mainland taxon A. × protomajoricum, of which haplotypes 2 and 4 were shared with the local populations of A. petrarchae subsp. bivalens. Haplotype 4 was the most common in both taxa (57 % in A. petrarchae subsp. bivalens and 67 % in A. × protomajoricum), while the closely related haplotypes 8 and 9 were restricted to A. × protomajoricum. Two haplotypes were present in A. majoricum. The Valencian population was invariant for haplotype 2, which was not detected among the A. petrarchae subsp. bivalens or A. × protomajoricum samples from this site. However, haplotype 2 was detected among samples of both these taxa from other sites in the Valencian region. The Majorcan populations of A. majoricum were invariant for haplotype 7, which was not detected from among the mainland populations, but is closely related to haplotypes 4 and 5, which were only detected among the Valencian samples of A. petrachae subsp. bivalens.
Fig. 4.
Minimum-spanning network of trnL-F chloroplast haplotypes in A. petrarchae subsp. bivalens, A. × protomajoricum and A. majoricum (defined in Table 1) and their geographical distribution among samples in these three taxa. See Table 2 for key to halotype number.
DISCUSSION
The allozyme phenotypes observed in A. × protomajoricum and A. majoricum in this study represent convincing additive banding patterns of alleles in the diploid species A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. We were thus able to identify unambiguously the alleles donated by the diploid parents for the vast majority of phenotypes in the hybrids and allotetraploids. Some observed phenotypes at four loci in A. majoricum are most plausibly explained by a null allele in one of the two homeologous genomes. Null alleles commonly arise by gene silencing following polyploidization (Werth and Windham, 1991; Soltis et al., 2004). The failure to observe the AAT-1 ‘84’ allele in A. fontanum subsp. fontanum which was picked up in A. × protomajoricum may simply reflect the high level of missing data (22·8 %) for this enzyme system among Valencian samples of A. fontanum subsp. fontanum.
Our molecular evidence corroborates previous cytological data (Sleep, 1967; Lovis and Reichstein, 1969; Pangua et al., 1992; Pérez Carro and Fernández Areces, 1992) demonstrating that A. × protomajoricum and A. majoricum are the diploid hybrid and allotetraploid, respectively, resulting from crosses between these two diploid parents. Such hybridization is a recurrent phenomenon in the genus Asplenium: many naturally occurring interspecific and intercytotypic hybrids have been described from Europe and elsewhere (Reichstein, 1981; Ranker et al., 1994; Matsumoto et al., 2003; Wang et al., 2003; Gabancho et al., 2010), and many of these have given rise to new polyploid species (Sleep, 1983; Vogel et al., 1999a).
Allotetraploids may arise either through the fusion of two diploid gametes, or via generation of a ‘triploid bridge’. In flowering plants, the triploid bridge pathway may play a significant role in the formation or maintenance of tetraploid populations (Ramsey and Schemske 1998; Husband 2004). However, it is more likely that A. majoricum arose directly from the diploid hybrid A. × protomajoricum, i.e. by the former route. First, in contrast to angiosperms, the production of two diploid gametes in homosporous ferns requires only a single unreduced meiosis in a diploid hybrid to yield a diplospore. The intragametophytic selfing system, unique to homosporous ferns, then offers a high probability that a (diploid) gametophyte grown from this diplospore will produce a tetraploid sporophyte. Lovis (reported in Reichstein, 1981) synthesized diploid hybrids between A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens which spontaneously yielded diplospores that gave rise to A. majoricum under experimental conditions, supporting the idea of direct chromosome doubling from A. × protomajoricum. Moreover, the vast majority of A. majoricum plants we analysed showed isozyme banding patterns consistent with balanced inheritance of alleles from one individual of each of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. Tetraploid formation via a triploid bridge would be more likely to result in unbalanced banding patterns.
The high frequency of hybridization among homosporous ferns has been explained by their relative lack of pre-zygotic reproductive isolating mechanisms (Schneller, 1981). Gender bias in hybridizations has been demonstrated in Asplenium × alternifolium (Vogel et al., 1998b) and Dryopteris × triploidea (Xiang et al., 2000). Our chloroplast DNA data show that strong gender bias is also characteristic of the hybridization between A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens: all A. × protomajoricum and A. majoricum analysed had A. petrarchae subsp. bivalens as the maternal parent. Like most other European Asplenium hybrids studied (Reichstein, 1981), A. × protomajoricum is largely sterile: the production of diplospores that can give rise to A. majoricum lineages occurs at very low frequency (Pangua et al., 1992; Perez Carro and Fernandez Areces, 1992). Viable spores were not observed on any of the individuals sampled, although we did not dissect all sporangia. The breeding system of homosporous ferns means that diploid hybrids will not undergo a gradual increase in fertility over several generations. The viable spores produced by such hybrids are unreduced diplospores, which then produce diploid gametophytes and gametes as discussed above. For this reason, and on the basis of the variation observed in isozyme phenotype patterns, we can be confident that almost all the A. × protomajoricum and at least some of the A. majoricum represent independent hybridization events. Hence, the observed unidirectionality of hybridization is highly statistically significant: given a null hypothesis in which the chances of chloroplast inheritance are 0·5 from each parent, the binomial probability of observing inheritance from only A. petrarchae subsp. bivalens in the 24 samples of A. × protomajoricum sequenced is 5·96 × 10−8. The mechanisms that might account for unidirectional hybridization in ferns are poorly understood. Vogel et al. (1998b) postulated that breeding system differences in the cross A. septentrionale × A. trichomanes favoured the outbreeding A. septentrionale as the maternal parent, accounting for the observed gender bias in the resulting hybrid A. × alternifolium. However, like most diploid fern species (Soltis et al., 1988), A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens are both largely outbreeding (Hunt et al., 2009).
Molecular analyses have established that multiple de novo origins of polyploid species are the rule rather than the exception (Soltis and Soltis, 1999; Soltis et al., 2004), and this is also true of homosporous ferns (Werth et al., 1985; Vogel et al., 1999b; Trewick et al., 2002; Perrie et al., 2010). From our allozyme data, it appears that there were at least four independent origins among the A. majoricum plants sampled in this study, based on the number of distinct phenotypes at the locus AAT-1. One alternative mechanism that could account for the observed multiple phenotypes, namely a single polyploidization event followed by introgression of alleles via backcrossing with diploid parents (Husband, 2004), seems unlikely in the case of A. majoricum. Several individuals of the triploid hybrid between A. majoricum and A. fontanum subsp. fontanum, A. × reichsteinii, have been reported from Majorca (Bennert et al., 1987), and one from Valencia (Pérez Carro and Fernández Areces, 1996). However, A. × reichsteinii is typically highly sterile and therefore unlikely to have played a significant role as a ‘triploid bridge’. Hybrids between A. majoricum and A. petrarchae subsp. bivalens are not known. Another potential source of alleles in A. majoricum is via introgression from autotetraploid cytotypes of the diploid parental taxa, produced via unreduced diplospores. The autotetraploid cytotype of A. petrarchae, A. petrarchae subsp. petrarchae, coexists with A. majoricum in both the Valencian and Majorcan populations of the latter. Rare tetraploid hybrids are known between A. majoricum and A. petrarchae subsp. petrarchae (A. × sollerense) from both regions (Lovis et al., 1969; Pérez Carro and Fernández Areces, 1996). However, A. × sollerense is highly sterile, with abortive spores and cytological examination showing a mixture of univalents, bivalents and trivalents at meiosis. Autotetraploid cytotypes of A. fontanum have never been found, although it is possible that initial-generation diploid gametophytes could exist. Although we cannot exclude a role for introgression, we suggest that most of the observed allozyme diversity in the allotetraploid arose directly from chromosome doubling in the diploid hybrid. This is supported by the sharing of additive banding patterns between A. × protomajoricum and A. majoricum, and the fact that these patterns had balanced gene dosages (in fixed heterozygotes) in a large majority of A. majoricum individuals. The minimum number of four independent allotetraploid origins would require that most of the MLGs observed have arisen through crossing between genetically distinct gametophytes in nascent A. majoricum populations.
All four AAT-1 compound alleles, and 13 distinct MLGs, are found in the A. majoricum population at Barranc de las Cuevas, indicating recurrent de novo polyploidization events within this small population. The A. majoricum MLGs found in Valencia today are genetically similar to a small number of the extant A. × protomajoricum individuals, while the remaining A. × protomajoricum MLGs form a cluster which has not given rise to any A. majoricum lineages (Fig. 1A). This cluster is differentiated by the allozyme phenotypes present at a single locus, 6-PGD (Supplementary Data Table S3). It is not clear whether the absence of A. majoricum individuals arising from these A. × protomajoricum MLGs reflects chance sampling of alleles in time and space, or whether some genotypes might be more likely than others to found locally successful polyploid lineages. Analysis of additional, more diverse loci from samples collected over a longer time period would be needed to resolve this question.
Multiple polyploid origins have been reported from many plant taxa, on varying geographical scales: they have also been found within a single population for Draba norvegica (Brochmann and Elven, 1992) and Heuchera grossulariifolia (Segraves et al., 1999). The small size of the Barranc de las Cuevas population and the large number of distinct allozyme genotypes suggest that recurrent de novo formation and extinction of polyploid individuals is occurring. However, the diversity of allozyme MLGs must be reconciled with the observation that all A. majoricum samples from Barranc de las Cuevas were monomorphic for a single chloroplast haplotype. This haplotype was found among A. petrarchae subsp. bivalens in Valencia as a whole, but not in this taxon or in A. × protomajoricum at Barranc de las Cuevas, suggesting it is locally rare and hence unlikely that multiple independent polyploidization events have captured the same haplotype by chance. It may be that there has been limited intergametophytic crossing in the nascent A. majoricum population here, such that a single maternal lineage has survived but alleles from multiple independent polyploidization events have been retained and shuffled by random assortment to give high nuclear diversity.
The opportunity for hybridization and polyploid formation is limited to a few localities, where it appears that the number of hybrids and polyploids fluctuates stochastically. At one site, Vallada, where large populations of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens co-occur, and where A. majoricum was reported (Pérez Carro and Fernández Areces, 1992), we did not find any hybrid or allotetraploid individuals. At two of the type sites for A. × protomajoricum, La Gola and Simat de la Valldigna, we found diploid hybrids growing together with both parental species, as reported by these authors (Pérez Carro and Fernández Areces, 1992), but no allotetraploids. Moreover, at Barranc de las Cuevas, where we carried out repeated sampling, we found little overlap of MLGs between individuals of A. majoricum sampled 3 years apart. This suggests that turnover of individuals may be rapid. The longevity of rock fern sporophytes is not well known but has been suggested to be 5–20 years (Ranker et al., 1994), although a study by Holderegger and Schneller (1994) indicated that individuals of A. septentrionale in small, isolated populations may be at least 30 years old. Further repeat monitoring of the population at Barranc de las Cuevas and other sites from which hybrids and polyploids in the A. majoricum complex have been recorded, involving precise mapping of individual plants, is needed to clarify the dynamics of plant turnover. However, the small number of A. majoricum individuals at Barranc de las Cuevas, and their high allozyme genetic diversity in comparison with the Majorcan populations (discussed below), suggest that the newly formed polyploids are struggling to become established long term at this site. The absence of A. majoricum individuals at other Valencian sites where A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens grow sympatrically corroborates this. Successful reproduction of A. majoricum is likely to be limited by strong competition for available niches at this and other sites. We observed that virtually all suitable horizontal and vertical fissures in the limestone rock face are densely populated by individuals of these and sometimes also other Asplenium taxa. Ecological competition is likely to operate in conjunction with minority cytotype exclusion, such that the much greater density of the outbreeding diploid parents at the site would result in back-crossing between A. majoricum and its diploid parents. This back-crossing would produce sterile triploid hybrids which constitute evolutionary dead-ends. These have been observed very rarely in nature: the sterile triploid A. × reichsteinii (A. majoricum × A. fontanum subsp. fontanum) has been recorded from Majorca (Bennert et al., 1987). Hybrids between A. majoricum and A. petrarchae subsp. bivalens have not been recorded in nature. We did not find any individuals at any of the sites sampled whose morphology or allozyme banding patterns suggested that they were likely to be triploid.
Prior to the discovery of the small Valencian populations of A. majoricum, this allotetraploid species was thought to be endemic to Majorca, where it occurs in much greater numbers. Our molecular data suggest that, in fact, the Majorcan populations themselves may also have their origin in mainland Spain rather than in situ on Majorca. This is supported by a comparison of the allele frequencies in A. majoricum with those in Majorcan and Valencian populations of A. petrarchae subsp. bivalens at the allozyme loci AAT-1 and PGM-2, and at the chloroplast trnL-F locus. Valencian populations of A. petrarchae subsp. bivalens include the alleles or haplotypes at these loci which are found at high frequency in Majorcan A. majoricum, whereas populations of A. petrarchae subsp. bivalens from Majorca are dominated by distinct alleles that are not found in the local allotetraploid populations. We cannot exclude the possibility that the few and small populations of A. petrarchae subsp. bivalens on Majorca are remnants of a former more extensive and genetically diverse population on the island, that included the alleles donated to A. majoricum. However, our evidence favours the hypothesis that A. majoricum did not evolve locally but colonized Majorca from the mainland. The alleles in A. majoricum donated by A. fontanum subsp. fontanum are also characteristic of those in the Valencian populations of the latter. However, since we do not know the genetic identity of the postulated now-extinct or near-extinct populations of A. fontanum subsp. fontanum on Majorca, this observation is not definitive with regard to the geographic origin of A. majoricum populations on the island.
The populations of A. majoricum on Majorca are genetically differentiated from those on Valencia, as can be seen by the network of their MLGs. The key locus accounting for this differentiation is 6-PGD, where all Majorcan samples have a genotype that we infer represents a null allele in the genome donated from A. fontanum subsp. fontanum (Supplementary Data Table S2). This is the only genetic change on the branch of the network separating the Majorcan MLG M14 from the Valencian MLG M3 (Fig. 1A). Together with the observation that null alleles from one or other parental genome characterize compound alleles specific to Majorcan A. majoricum at three other loci (MDH-1, PGM-2 and SKDH), this minor genetic change is consistent with the interpretation that these genotypes evolved from closely related Valencian genotypes by a process of gene silencing. The increased frequency of null alleles in the Majorcan populations probably reflects their longer evolutionary history relative to recurrently formed A. majoricum in Valencia.
Homosporous ferns disperse easily via wind-borne spores. The allotetraploid A. adiantum-nigrum has colonized Hawaii from mainland North America on at least three occasions (Ranker et al., 1994), and A. hookerianum appears to have colonized the Chatham Islands from New Zealand at least twice (Shepherd et al., 2009). In both the above cases, multiple source populations exist on the mainland, in contrast to the case of A. majoricum. However, notwithstanding the much greater geographical scale over which dispersal has occurred in North American A. adiantum-nigrum, there is some similarity with the dispersal of A. majoricum: mainland populations are rare in comparison with the island populations to which they have given rise. Our data show that despite the large population size, genetic diversity in A. majoricum is very low. A single MLG makes up the vast majority of samples, and most other MLGs differ from this one by the presence of null alleles, which are likely to result from the silencing of duplicated genes (Werth and Windham, 1991). The Majorcan populations of A. majoricum could have originated from a single colonization event from Valencia, followed by reproduction through intragametophytic selfing. Vogel et al. (1999b) have argued that a switch from outbreeding diploids to inbreeding polyploids occurs in multiple European Asplenium taxa, with the deleterious effects of inbreeding buffered by the fixed heterozygosity of the polyploid genome, and that this has facilitated the range expansion of polyploid taxa in colonizing new habitat. We can speculate that this mechanism has also enabled the establishment of the large Majorcan populations of A. majoricum. However, the allozyme markers we used were not sufficiently variable to give statistically meaningful FIS estimates for the inference of breeding system.
The contrast in the success of establishment of the Majorcan and Valencian populations of A. majoricum is notable. The success of the Majorcan population cannot, however, be attributed to greater genetic diversity. In a similar result, Abbott et al. (2007) found that populations of the neopolyploid Senecio cambrensis in Wales are declining despite high levels of diversity. The A. majoricum population in Valencia occurs on a natural limestone outcrop, where the plants grow together with their diploid progenitors. In contrast, the Majorcan populations largely grow on artificial limestone walls surrounding olive terraces. According to our observations, neither A. petrarchae subsp. bivalens nor A. fontanum subsp. fontanum grow in this habitat today, although the presence of the hybrid A. × reichsteinii indicates that A. fontanum subsp. fontanum, or at least its gametophytes, has grown there in the recent past (Bennert et al., 1990). In the course of this study we discovered a new, smaller, Majorcan population of A. majoricum at Caimari (MAJ-4), which grows on natural limestone outcrop together with A. petrarchae subsp. bivalens. However, the A. majoricum plants here are genetically uniform with those in the other populations on Majorca (Supplementary Data Table S3), and do not appear to be descended from the local A. petrarchae subsp. bivalens.
We hypothesize that the chance founding of A. majoricum populations in the limestone wall habitat where neither A. fontanum subsp. fontanum nor A. petrarchae subsp. bivalens occurs has been a key factor that has enabled the establishment of the allotetraploid species. This resonates with the situation in other diploid–polyploid complexes in European Asplenium, although the habitat for A. majoricum has been created by anthropogenic activity rather than the glacial cycles discussed by Vogel et al. (1999b). However, these different processes achieve the same result: they free polyploid taxa from competition with their diploid progenitors. An alternative hypothesis is that the ecological niche of this habitat shift is more favourable to A. majoricum, and it has outcompeted existing populations of A. fontanum subsp. fontanum and A. petrarchae subsp. bivalens. However, if these diploids coexisted locally, it would be expected that populations of A. majoricum would arise indigenously. In this case, as discussed above, an explanation would be needed for the discrepancy in alleles and haplotypes between Majorcan A. petrarchae subsp. bivalens and A. majoricum. The most parsimonious explanation in the light of the available data therefore appears to be the colonization of vacant habitat by extraneously originating A. majoricum, but further work on the fine-scale ecological preferences of these taxa would be revealing with regard to the likely outcome of interspecific competition between them. Common-garden experiments would also be valuable to determine whether selection has resulted in local adaptation in the disjunct A. majoricum populations.
Conclusions
Our data provide new understanding on the origins of A. majoricum and its history in the fern flora of the Balearic islands. The likely source of these populations in mainland Spain demonstrates the evolutionary significance of extremely rare and isolated occurrences of hybrids and polyploids, a topic with rich potential for further exploration among the European Asplenium flora. In the A. majoricum species complex, it appears that ecological opportunity has been more important than genetic factors in facilitating the establishment of polyploid populations. This species complex provides an excellent system for further long-term monitoring and analysis using new molecular tools, which will provide finer resolution of the interplay between ecological, genetic and breeding system factors in determining polyploid success in natural populations.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the late Professor Michael Majerus for advice, Michael Grundmann and Cecília Durães for help in the lab, Fred Rumsey and Antonia Eastwood for help collecting samples, and Cameron Petrie for assistance with graphics. This work was supported by a Natural Environment Research Council PhD studentship and grants from St John's College, Cambridge, and the Department of Genetics, University of Cambridge to H.V.H.
LITERATURE CITED
- Abbott RJ, Ireland HE, Rogers HJ. Population decline despite high genetic diversity in the new allopolyploid species Senecio cambrensis (Asteraceae) Molecular Ecology. 2007;16:1023–1033. doi: 10.1111/j.1365-294X.2007.03169.x. [DOI] [PubMed] [Google Scholar]
- Barringer BC. Polyploidy and self-fertilization in flowering plants. American Journal of Botany. 2007;94:1527–1533. doi: 10.3732/ajb.94.9.1527. [DOI] [PubMed] [Google Scholar]
- Bennert HW, Rasbach H, Rasbach K. Asplenium × reichsteinii (= Asplenium fontanum × A. majoricum; Aspleniaceae: Pteridophyta), a new endemic fern hybrid from Mallorca, Balearic Islands. Fern Gazette. 1987;13:133–141. [Google Scholar]
- Bennert HW, Rasbach H, Rasbach K. Asplenium petrarchae (Guérin) DC. subsp. bivalens und Asplenium × helii nothosubsp. calobrense – Neufunde auf der Insel Mallorca. Farnblätter. 1990;21:15–26. [Google Scholar]
- Brochmann C, Elven R. Ecological and genetic consequences of polyploidy in Arctic Draba (Brassicaceae) Evolutionary Trends in Plants. 1992;6:111–124. [Google Scholar]
- Coyne JA, Orr HA. Speciation. New York: W.H. Freeman; 2004. [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mousadik A, Petit RJ. High level of genetic differentiationfor allelic richness among populations of the Argan tree (Argania spinosa (L.) Skeels) endemic to Morocco. Theoretical and Applied Genetics. 1996;92:832–839. doi: 10.1007/BF00221895. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3·0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Gabancho LR, Prada C, Galan JMGY. Sexuality and apogamy in the Cuban Asplenium auritum-monodon complex (Aspleniaceae) Plant Systematics and Evolution. 2010;289:137–146. [Google Scholar]
- Goudet J. FSTAT (vers. 1·2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. Version 2·9·3 available from http://www.unil.ch/izea/softwares/fstat.html . [Google Scholar]
- Guggisberg A, Mansion G, Kelso S, Conti E. Evolution of biogeographic patterns, ploidy levels, and breeding systems in a diploid–polyploid species complex of Primula. New Phytologist. 2006;171:617–632. doi: 10.1111/j.1469-8137.2006.01722.x. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Hiscock SJ. Genomic clues to the evolutionary success of polyploid plants. Current Biology. 2008;18:R435–R444. doi: 10.1016/j.cub.2008.03.043. [DOI] [PubMed] [Google Scholar]
- Herrero Nieto A, Prada Moral C. Nuevas citas para tres híbridos de Asplenium (Aspleniaceae, Pteridophyta) en la Península Ibérica. Acta Botanica Malacitana. 1997;22:19–27. [Google Scholar]
- Holderegger R, Schneller JJ. Are small isolated populations of Asplenium septentrionale variable? Biological Journal of the Linnean Society. 1994;51:377–385. [Google Scholar]
- Hunt HV. Evolution in the Asplenium majoricum complex. 2004. PhD thesis, University of Cambridge, UK. [Google Scholar]
- Hunt HV, Ansell SW, Russell SJ, Schneider H, Vogel JC. Genetic diversity and phylogeography in two diploid ferns, Asplenium fontanum subsp. fontanum and A. petrarchae subsp. bivalens, in the western Mediterranean. Molecular Ecology. 2009;18:4940–4954. doi: 10.1111/j.1365-294X.2009.04402.x. [DOI] [PubMed] [Google Scholar]
- Husband BC. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society. 2004;82:537–546. [Google Scholar]
- Husband BC, Ozimec B, Martin SL, Pollock L. Mating consequences of polyploid evolution in flowering plants: current trends and insights from synthetic polyploids. International Journal of Plant Sciences. 2008;169:195–206. [Google Scholar]
- Huson DH, Richter DC, Rausch C, et al. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. doi:10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibars AM, Herrero-Borgoñon JJ, Estrelles E, Martínez I. Helechos de la Comunidad Valenciana. Valencia: Generalitat Valenciana, Conselleria de Medio Ambiente; 1999. [Google Scholar]
- Jacquotot MC, Orell J. Asplenium majoricum R. Litardière, su área de expansión en la sierra norte de Mallorca. Collectanea Botanica. 1968;7:559–572. [Google Scholar]
- Kephart SR. Starch gel electrophoresis of plant isozymes: a comparative analysis of techniques. American Journal of Botany. 1990;77:693–712. [Google Scholar]
- Levin D. Minority cytotype exclusion in local plant populations. Taxon. 1975;24:35–43. [Google Scholar]
- Levin D. The role of chromosomal change in plant evolution. Oxford: Oxford University Press; 2002. [Google Scholar]
- Lewis PO, Zaykin D. Genetic data analysis: computer program for the analysis of allelic data. Version 1·0 (d16c) 2001 http://lewis.eeb.uconn.edu/lewishome/software.html . [Google Scholar]
- Litardière R. Contribution à l'étude de la flora ptéridologique de la péninsule ibérique. Bulletin de Géographie Botanique. 1911;21:28–29. [Google Scholar]
- Liu K, Muse SV. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lovis JD, Reichstein T. Der Farnbastard Asplenium × orellii hybr. nov.=Asplenium majoricum Litard. × A. trichomanes L. subsp. quadrivalens D.E. Meyer und die Abstammung von A. majoricum. Berichte der Schweizerischen Botanischen Gesellschaft. 1969;79:335–345. [Google Scholar]
- Lovis JD, Sleep A, Reichstein T. Der Farnbastard Asplenium × sollerense hybr. nov.=Asplenium majoricum Litard. × A. petrarchae (Guérin) DC. subsp. petrarchae. Berichte der Schweizerischen Botanischen Gesellschaft. 1969;79:369–375. [Google Scholar]
- Matsumoto S, Iwashina T, Kitajima J, Mitsuta S. Evidence by flavonoid markers of four natural hybrids among Asplenium normale and related species (Aspleniaceae) in Japan. Biochemical Systematics and Ecology. 2003;31:51–58. [Google Scholar]
- Meimberg H, Rice KJ, Milan NF, Njoku CC, McKay JK. Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae) American Journal of Botany. 2009;96:1262–1273. doi: 10.3732/ajb.0800345. [DOI] [PubMed] [Google Scholar]
- Moran RC. The Asplenium trichomanes complex in the United States and adjacent Canada. American Fern Journal. 1982;72:5–11. [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. Journal of Molecular Evolution. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Pangua E, Prada C, Pajarón S, Salvo E. A new Asplenium hybrid from Valencia (Spain) related to A. majoricum Litard. Botanical Journal of the Linnean Society. 1992;108:1–13. [Google Scholar]
- Pérez Carro FJ, Fernández Areces MP. Asplenium × protomajoricum hybr. nov. (A. fontanum subsp. fontanum × A. petrarchae subsp. bivalens) y precisiones corológicas sobre A. majoricum en el Levante español. Anales Jardín Botánico de Madrid. 1992;49:187–194. [Google Scholar]
- Pérez Carro FJ, Fernández Areces MP. Híbridos del género Asplenium L. (Aspleniaceae) en la Península Ibérica. Anales Jardín Botánico de Madrid. 1996;54:106–125. [Google Scholar]
- Perrie LR, Shepherd LD, de Lange PJ, Brownsey PJ. Parallel polyploid speciation: distinct sympatric gene-pools of recurrently derived allo-octoploid Asplenium ferns. Molecular Ecology. 2010;19:2916–2932. doi: 10.1111/j.1365-294X.2010.04705.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, El-Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12:844–855. [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annual Review of Ecology and Systematics. 2002;33:589–639. [Google Scholar]
- Ranker TA, Floyd SK, Trapp PG. Multiple colonizations of Asplenium adiantum-nigrum onto the Hawaiian archipelago. Evolution. 1994;48:1364–1370. doi: 10.1111/j.1558-5646.1994.tb05319.x. [DOI] [PubMed] [Google Scholar]
- Reichstein T. Hybrids in European Aspleniaceae (Pteridophyta) Botanica Helvetica. 1981;91:89–139. [Google Scholar]
- Schneider H, Russell SJ, Cox CJ, et al. Chloroplast phylogeny of Asplenioid ferns based on rbcL and trnL–F spacer sequences (Polypodiidae, Aspleniaceae) and its implications for biogeography. Systematic Botany. 2004;29:260–274. [Google Scholar]
- Schneller J. Evidence for intergeneric incompatibility in ferns. Plant Systematics and Evolution. 1981;137:45–56. [Google Scholar]
- Segraves KA, Thompson JN, Soltis PS, Soltis DE. Multiple origins of polyploidy and the geographic structure of Heuchera grossulariifolia. Molecular Ecology. 1999;8:253–263. [Google Scholar]
- Shepherd LD, de Lange PJ, Perrie LR. Multiple colonizations of a remote oceanic archipelago by one species: how common is long-distance dispersal? Journal of Biogeography. 2009;36:1972–1977. [Google Scholar]
- Sleep A. A contribution to the cytotaxonomy of Asplenium majoricum. British Fern Gazette. 1967;9:321–329. [Google Scholar]
- Sleep A. On the genus Asplenium in the Iberian peninsula. Acta Botanica Malacitana. 1983;8:11–46. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology and Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Haufler CH, Darrow DC, Gastony GJ. Starch gel electrophoresis of ferns: a compilation of grinding buffers, gel and electrode buffers, and staining schedules. American Fern Journal. 1983;73:9–27. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since Plant Speciation. New Phytologist. 2004;161:173–191. [Google Scholar]
- Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences, USA. 2000;97:7051–7057. doi: 10.1073/pnas.97.13.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annual Review of Plant Biology. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Holsinger KE. Estimates of intragametophytic selfing and interpopulational gene flow in homosporous ferns. American Journal of Botany. 1988;75:1765–1770. [Google Scholar]
- Suter M, Schneller JJ, Vogel JC. Investigations into the genetic variation, population structure, and breeding systems of the fern Asplenium trichomanes subsp. quadrivalens. International Journal of Plant Sciences. 2000;161:233–244. doi: 10.1086/314258. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Trewick SA, Morgan-Richards M, Russell SJ, et al. Polyploidy, phylogeography and Pleistocene refugia of the rockfern Asplenium ceterach: evidence from chloroplast DNA. Molecular Ecology. 2002;11:2003–2012. doi: 10.1046/j.1365-294x.2002.01583.x. [DOI] [PubMed] [Google Scholar]
- Vogel JC, Russell SJ, Rumsey FJ, Barrett JA, Gibby M. Evidence for maternal transmission of chloroplast DNA in the genus Asplenium (Aspleniaceae, Pteridophyta) Botanica Acta. 1998a;111:247–249. [Google Scholar]
- Vogel JC, Russell SJ, Rumsey FJ, Barrett JA, Gibby M. On hybrid formation in the rock fern Asplenium × alternifolium (Aspleniaceae, Pteridophyta) Botanica Acta. 1998b;111:241–246. [Google Scholar]
- Vogel JC, Rumsey FJ, Schneller JJ, Barrett JA, Gibby M. Where are the glacial refugia in Europe? Evidence from pteridophytes. Biological Journal of the Linnean Society. 1999a;66:23–37. [Google Scholar]
- Vogel JC, Barrett JA, Rumsey FJ, Gibby M. Identifying multiple origins in homosporous pteridophytes. In: Hollingsworth PM, Bateman RM, Gornall RJ, editors. Molecular Systematics and Plant Evolution. London: Taylor & Francis; 1999b. pp. 101–117. [Google Scholar]
- Wagner WH, Wagner FS, Taylor WC. Detecting abortive spores in herbarium specimens of sterile hybrids. American Fern Journal. 1985;76:129–140. [Google Scholar]
- Wang ZR, Wang KQ, Zhang F, Hou X. A biosystematic study on Asplenium sarelii complex. Acta Botanica Sinica. 2003;45:1–14. [Google Scholar]
- Wendel JF, Weeden NF. Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS, editors. Isozymes in plant biology. Portland, OR: Dioscorides Press; 1989. pp. 5–45. [Google Scholar]
- Werth CR, Windham MD. A model for divergent, allopatric speciation of polyploid pteridophytes resulting from silencing of duplicate gene expression. American Naturalist. 1991;137:515–526. [Google Scholar]
- Werth CR, Guttman SI, Eshbaugh WH. Recurring origins of allopolyploid species in Asplenium. Science. 1985;228:731–733. doi: 10.1126/science.228.4700.731. [DOI] [PubMed] [Google Scholar]
- Whitton J. One down and thousands to go – dissecting polyploid speciation. New Phytologist. 2004;161:607–610. doi: 10.1111/j.1469-8137.2003.01017.x. [DOI] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, et al. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and genetics of populations, Volume 4. Variability within and among natural populations. Chicago: University of Chicago Press; 1978. [Google Scholar]
- Xiang L, Werth CR, Emery SN, McCauley DE. Population-specific gender-biased hybridization between Dryopteris intermedia and D. carthusiana: evidence from chloroplast DNA. American Journal of Botany. 2000;87:1175–1180. [PubMed] [Google Scholar]
- Yeh FC, Yang R, Boyle T. POPGENE. Microsoft Window based freeware for population genetic analysis. 1999 Version 1·32. Centre for International Forestry Research, University of Alberta. Available at http://www.ualberta.ca/~fyeh/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.